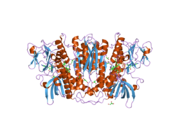User:Zwickipedia/sandbox
The flavoenzyme glutathione reductase (EC 1.8.1.7), also known as GSR or GR, catalyzes the reduction of glutathione disulfide (GSSG) to the sulfhydryl form glutathione (GSH), which is a critical molecule in resisting oxidative stress and maintaining the reducing environment of the cell.[1][2][3] Glutathione reductase functions as dimeric disulfide oxidoreductase and utilizes an FAD prosthetic group and NADPH to reduce one mole of GSSG to two moles of GSH, as shown in Scheme 1.
The glutathione reductase is conserved between all kingdoms. In bacteria, yeasts, and animals, one glutathione reductase gene is found; however, in plant genomes, two GR genes are encoded. Drosophila and Trypanosomes do not have any GR at all.[4] In these organisms, glutathione reduction is performed by either the thioredoxin or the trypanothione system, respectively.[4][5]
History
[edit]Glutathione reductase was first purified in 1955 at Yale by E. Racker.[6] Racker also identified NADPH as the primary electron donor for the enzyme. Later groups confirmed the presence of FAD and the thiol group, and an initial mechanism was suggested for the mechanism in 1965.[7][8] The initial (low resolution) structure of glutathione reductase was solved in 1977. This was quickly followed by a 3Å structure by Shulze et al in 1978.[9] Glutathione reductase has been studied exhaustively since these early experiments, and is subsequently one of the most well characterized enzymes to date.
Function
[edit]Glutathione play a key role in maintaining proper function and preventing oxidative stress in human cells. It can act as a scavanger for hydroxyl radicals,singlet oxygens, and other various electrophiles. In addition, it plays a key role in the metabolism and clearance of xenobiotics, acts as a cofactor in ceratin detoxifying enzymes, participates in transport, and regenerates antioxidants such and Vitamins E and C to their reactive forms. The ratio of GSSH/GSH present in the cell is a key factor in properly maintaining the oxidative balance of the cell, that is, it is critical that the cell maintains high levels of the reduced glutathione and a low level of the oxidized Glutathione disulfide. This narrow balance is maintained by glutathione reductase, which catalyzes the reduction of GSSG to GSH.[1]
Structure
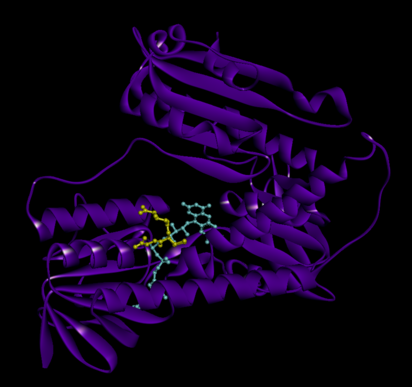
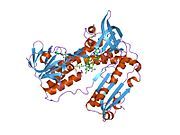
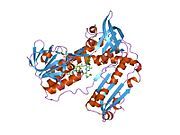
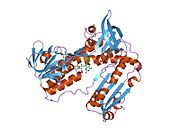
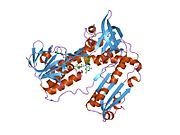
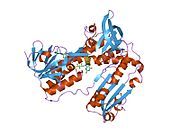
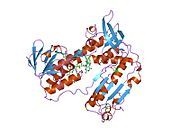
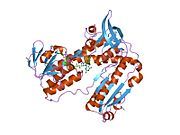
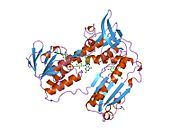
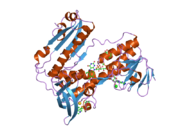
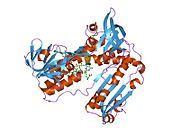
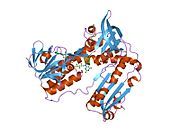
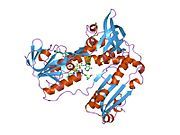
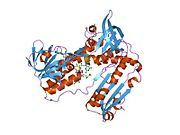
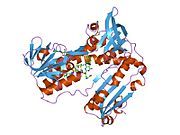
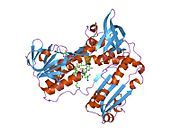
[edit]Glutathione reductase from human erythrocytes is a homodimer consisting of 52Kd monomers, each containing 3 domains. GR exhibits single sheet, double layered topology where an anti-parallel beta-sheet is largely exposed to the solvent on one face while being covered by random coils on the other face.[10] This includes and NADPH-binding Domain, FAD-binding domain(s) and a dimerization domain. Each monomer contains 478 residues and one FAD molecule. GR is a thermostable protein, retaining function up to 65oC.[11][12]
Reaction Mechanism
[edit]Steps:
1) NADPH binding to the oxidized enzyme
2) Reduction of FAD to FADH- anion by NADPH
3) Reduced FADH- anion collapses into a charge relay complex and reduces Cys58-Cys63 disulfide
4) Oxidized Glutathione disulfide binds to the reduced enzyme and forms a mixed disulfide with Cys58 and releases one reduced glutathione
5) Cys63 attacks the mixed disulfide on Cys58 to release a reduced glutathione and reform the redox active disulfide
Reductive Half
[edit]The action of GR proceeds through two distinct half reactions, a reductive half mechanism followed by an oxidative half. In the first half, NADPH reduces FAD present in GSR to produce a transient FADH- anion. This anion then quickly breaks a disulfide bond of Cys58 - Cys63, forming a short lived covalent bond a stable charge-transfer complex between the flavin and Cys63. The now oxidized NADP+ is released and is subsequently replaced by a new molecule of NADPH. This is the end of the so called reductive half of the mechanism.
Oxidative Half
[edit]In the oxidative half of the meachanism, Cys63 nucleophilically attacks the nearest sulfide unit in the GSSG molecule (promoted by His467), which creates a mixed disulfide bond (GS-Cys58) and a GS- anion. His467 of GSR then protonates the GS- anion to release the first molecule of GSH. Next, Cys58 nucleophilically attacks the sulfide of Cys58, releasing a GS- anion, which, in turn, picks up a solvent proton and is released from the enzyme, thereby creating the second GSH. So, for every GSSG and NADPH, two reduced GSH molecules are gained, which can again act as antioxidants scavenging reactive oxygen species in the cell.[13]
Medical Relevance of Glutathione reductase in human cells
[edit]GSH is a key cellular antioxidant and plays a major role in the phase 2 metabolic clearance of electrophilic xenobiotics. The importance of the GSH pathway and enzymes that affect this delicate balance is gaining an increased level of attention in recent years. Although glutathione reductase has been an attractive target for many pharmaceuticals, there have been no successful glutathione reductase related therapeutic compounds created to date. In particular, glutathione reductase appears to be a good target for anti-malarials, as the glutathione reductase of the malaria parasite p. falciparum has a significantly different protein fold than that of mammalian glutathione reductase.[14][9] By designing drugs specific to p. Falciparum it may be possible to selectively induce oxidative stress in the parasite, while not affecting the host.
There are two main classes of GR targeting compounds:[15][16][17][18]
- Inhibitors of GSSG binding, or dimerization: Reactive electrophiles such as gold compounds, and fluoronaphthoquinones.
- Drugs which use glutathione reductase to regenerate, such as redox cyclers. Two examples of these types of compounds are Methylene blue and Naphthoquinone.
Clinical trials performed in Burkina Faso have revealed mixed results when treating malaria with Naphthoquinones
In cells exposed to high levels of oxidative stress, like red blood cells, up to 10% of the glucose consumption may be directed to the pentose phosphate pathway (PPP) for production of the NADPH needed for this reaction. In the case of erythrocytes, if the PPP is non-functional, then the oxidative stress in the cell will lead to cell lysis and anemia.[19]
Glutathione reductase deficiency is a rare disorder in which the glutathione reductase activity is absent from erythrocytes, leukocytes or both. In one study this disorder was observed in only two cases in 15,000 tests for glutathione reductase deficiency performed over the course of 30 years.[20] In the same study, glutathione reductase deficiency was associated with cataracts and favism in one patient and their family, and with severe unconjugated hyperbilirubinemia in another patient.[20]
Glutathione reductase in plant cells
[edit]As it does in human cells, glutathione reductase helps to protect plant cells from reactive oxygen species. In plants, glutathione reductase contributes to plants' responses to abiotic stress, contributing to plants' ability to survive in dynamic environments.[21] Glutathione reductase activity has been shown to be modulated in response to metals, metalloids, salinity, drought, UV radiation and heat induced stress.[21]
Monitoring glutathione reductase activity
[edit]The activity of glutathione reductase is used as indicator for oxidative stress. The activity can be monitored by the NADPH consumption, with absorbance at 340 nm, or the formed GSH can be visualized by Ellman's reagent.[22] Alternatively the activity can be measured using roGFP (redox-sensitive Green Fluorescent Protein).[23]
Interactive pathway map
[edit]Interactive pathway can be found here: pathway map
References
[edit]- ^ a b Deponte, Marcel (May 2013). "Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes". Biochimica et Biophysica Acta (BBA) - General Subjects. 1830 (5): 3217–3266. doi:10.1016/j.bbagen.2012.09.018. PMID 23036594.
- ^ Meister, A (1988 Nov 25). "Glutathione metabolism and its selective modification". The Journal of Biological Chemistry. 263 (33): 17205–8. doi:10.1016/S0021-9258(19)77815-6. PMID 3053703.
{{cite journal}}: Check date values in:|date=(help) - ^ Mannervik, B (1987 Aug). "The enzymes of glutathione metabolism: an overview". Biochemical Society Transactions. 15 (4): 717–8. doi:10.1042/bst0150717. PMID 3315772.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Müller HM, Botella-Munoz J, Schneuwly S, Schirmer R, Becker K (2001). "Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster". Science. 291 (5504): 643–6. doi:10.1126/science.291.5504.643. PMID 11158675.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Krauth-Siegel RL, Comini MA (2008). "Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism". Biochim Biophys Acta. 1780 (11): 1236–48. doi:10.1016/j.bbagen.2008.03.006. PMID 18395526.
- ^ Racker, E. (1955-12-01). "Glutathione Reductase from Bakers' Yeast and Beef Liver". Journal of Biological Chemistry. 217 (2): 855–866. ISSN 1083-351X 0021-9258, 1083-351X. PMID 13271446. Retrieved 2014-02-04.
{{cite journal}}: Check|issn=value (help) - ^ Massey, V.; Williams Jr, C. H. (November 1965). "On the reaction mechanism of yeast glutathione reductase". The Journal of Biological Chemistry. 240 (11): 4470–4480. doi:10.1016/S0021-9258(18)97085-7. ISSN 0021-9258. PMID 4378936.
- ^ MAPSON LW; ISHERWOOD FA (January 1963). "Glutathione reductase from germinated peas". The Biochemical Journal. 86 (1): 173–191. doi:10.1042/bj0860173. ISSN 0264-6021. PMC 1201730. PMID 13932735.
- ^ a b Schulz, G. E.; Schirmer, R. H.; Sachsenheimer, W.; Pai, E. F. (1978-05-11). "The structure of the flavoenzyme glutathione reductase". Nature. 273 (5658): 120–124. doi:10.1038/273120a0. ISSN 0028-0836. PMID 25387.
- ^ Grisham, Reginald H. Garrett,... Charles M. (2005). Biochemistry (3rd ed.). Belmont, CA: Thomson Brooks/Cole. ISBN 0534490336.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Masella, R.; Di Benedetto, R.; Varì, R.; Filesi, C.; Giovannini, C. (2005 Oct). "Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes". The Journal of Nutritional Biochemistry. 16 (10): 577–86. doi:10.1016/j.jnutbio.2005.05.013. PMID 16111877.
{{cite journal}}: Check date values in:|date=(help) - ^ Dym, Orly; Eisenberg, David (September 2001). "Sequence-structure analysis of FAD-containing proteins". Protein Science. 10 (9): 1712–1728. doi:10.1110/ps.12801. PMC 2253189. PMID 11514662.
- ^ Berkholz, Donald S.; Faber, H. Richard; Savvides, Savvas N.; Karplus, P. Andrew (October 2008). "Catalytic Cycle of Human Glutathione Reductase Near 1 Å Resolution". Journal of Molecular Biology. 382 (2): 371–384. doi:10.1016/j.jmb.2008.06.083. PMC 2593804. PMID 18638483.
- ^ Sarma, G. N.; Savvides, S. N.; Becker, K.; Schirmer, M.; Schirmer, R. H.; Karplus, P. A. (2003-05-09). "Glutathione reductase of the malarial parasite Plasmodium falciparum: crystal structure and inhibitor development". Journal of Molecular Biology. 328 (4): 893–907. doi:10.1016/s0022-2836(03)00347-4. ISSN 0022-2836. PMID 12729762.
- ^ Buchholz, Kathrin; Schirmer, R. Heiner; Eubel, Jana K.; Akoachere, Monique B.; Dandekar, Thomas; Becker, Katja; Gromer, Stephan (January 2008). "Interactions of methylene blue with human disulfide reductases and their orthologues from Plasmodium falciparum". Antimicrobial Agents and Chemotherapy. 52 (1): 183–191. doi:10.1128/AAC.00773-07. ISSN 0066-4804. PMC 2223905. PMID 17967916.
- ^ Müller, Tobias; Johann, Laure; Jannack, Beate; Brückner, Margit; Lanfranchi, Don Antoine; Bauer, Holger; Sanchez, Cecilia; Yardley, Vanessa; Deregnaucourt, Christiane; Schrével, Joseph; Lanzer, Michael; Schirmer, R. Heiner; Davioud-Charvet, Elisabeth (2011-08-03). "Glutathione reductase-catalyzed cascade of redox reactions to bioactivate potent antimalarial 1,4-naphthoquinones--a new strategy to combat malarial parasites". Journal of the American Chemical Society. 133 (30): 11557–11571. doi:10.1021/ja201729z. ISSN 1520-5126. PMID 21682307.
- ^ Deponte, Marcel; Urig, Sabine; Arscott, L. David; Fritz-Wolf, Karin; Réau, Régis; Herold-Mende, Christel; Koncarevic, Sasa; Meyer, Markus; Davioud-Charvet, Elisabeth; Ballou, David P.; Williams, Charles H.; Becker, Katja (2005-05-27). "Mechanistic studies on a novel, highly potent gold-phosphole inhibitor of human glutathione reductase". The Journal of Biological Chemistry. 280 (21): 20628–20637. doi:10.1074/jbc.M412519200. ISSN 0021-9258. PMID 15792952.
- ^ Deponte, Marcel (May 2013). "Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes". Biochimica et Biophysica Acta (BBA) - General Subjects. 1830 (5): 3217–3266. doi:10.1016/j.bbagen.2012.09.018. ISSN 0006-3002. PMID 23036594.
- ^ Champe, et al. Biochemistry, Fourth Edition. Lippincott Williams and Wilkins. 2008
- ^ a b Kamerbeek NM, Zwieten R, Boer M, Morren G, Vuil H, Bannink N, Lincke C, Dolman KM, Becker K, Schirmer RH, Gromer S, Roos D (2007). "Molecular basis of glutathione reductase deficiency in human blood cells". Blood. 109 (8): 3560–3566. doi:10.1182/blood-2006-08-042531. PMID 17185460.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Gill, Sarvajeet Singh; Anjum, Naser A.; Hasanuzzaman, Mirza; Gill, Ritu; Trivedi, Dipesh Kumar; Ahmad, Iqbal; Pereira, Eduarda; Tuteja, Narendra (2013). "Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations". Plant Physiology and Biochemistry. 70: 204–212. doi:10.1016/j.plaphy.2013.05.032. PMID 23792825.
- ^ Smith IK, Vierheller TL, Thorne CA (1988). "RAssay of glutathione reductase in crude tissue homogenates using 5,5'-dithiobis(2-nitrobenzoic acid)". Anal Biochem. 175 (2): 408–13. doi:10.1016/0003-2697(88)90564-7. PMID 3239770.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R. (2009). "The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis". Proc Natl Acad Sci U S A. 106 (22): 9109–14. doi:10.1073/pnas.0900206106. PMC 2690020. PMID 19451637.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Further reading
[edit]Category:Antioxidants Category:EC 1.8.1