User:Jjcantu/Original
| amidophosphoribosyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.4.2.14 | ||||||||
| CAS no. | 9031-82-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme that in humans is encoded by the PPAT (phosphoribosyl pyrophosphate amidotransferase) gene.[1][2]
Function
[edit]ATase is an enzyme that converts α-phosphoribosylpyrophosphate (α-PRPP) into 5-β-phosphoribosylamine. The enzyme uses the ammonia group from the glutamine side-chain. This is the committing step in de novo purine synthesis. It is allosterically inhibited by AMP, GMP, and IMP. 6TGMP (6-thioguanine monophosphate) is a pseudo inhibitor for ATase
ATase is a member of the purine/pyrimidine phosphoribosyltransferase family. This protein is a regulatory allosteric enzyme that catalyzes the first step of de novo purine nucleotide biosynthesis.[1]
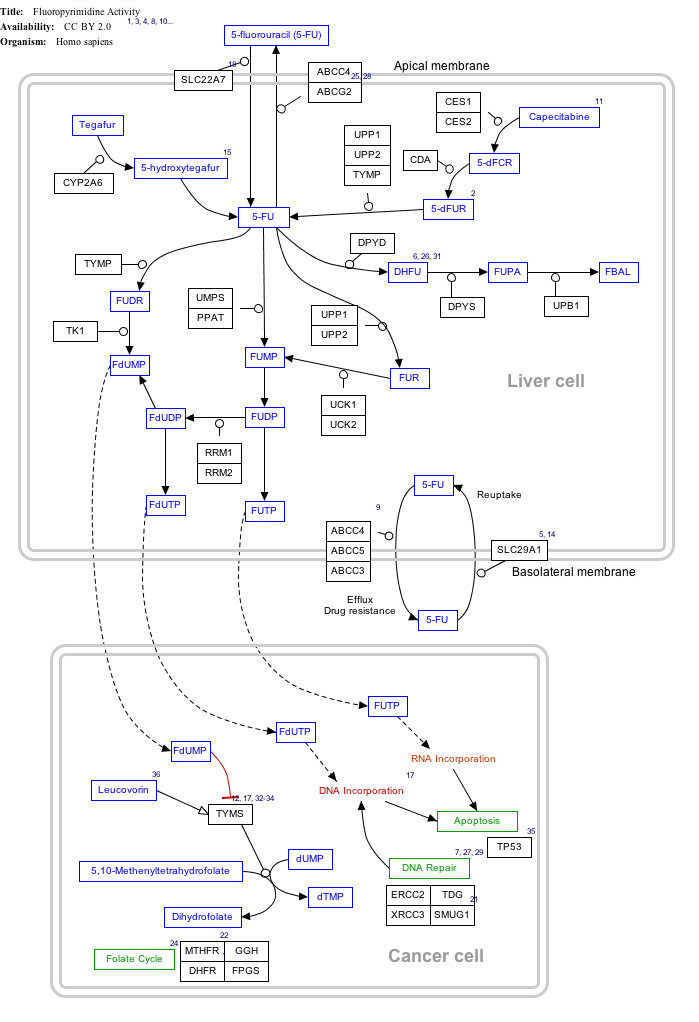
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "FluoropyrimidineActivity_WP1601".
References
[edit]- ^ a b "Entrez Gene: phosphoribosyl pyrophosphate amidotransferase".
- ^ Brayton KA, Chen Z, Zhou G, Nagy PL, Gavalas A, Trent JM, Deaven LL, Dixon JE, Zalkin H (Feb 1994). "Two genes for de novo purine nucleotide synthesis on human chromosome 4 are closely linked and divergently transcribed". The Journal of Biological Chemistry. 269 (7): 5313–21. doi:10.1016/S0021-9258(17)37689-5. PMID 8106516.
Further reading
[edit]- Iwahana H, Oka J, Mizusawa N, Kudo E, Ii S, Yoshimoto K, Holmes EW, Itakura M (Jan 1993). "Molecular cloning of human amidophosphoribosyltransferase". Biochemical and Biophysical Research Communications. 190 (1): 192–200. doi:10.1006/bbrc.1993.1030. PMID 8380692.
- Gassmann MG, Stanzel A, Werner S (Nov 1999). "Growth factor-regulated expression of enzymes involved in nucleotide biosynthesis: a novel mechanism of growth factor action". Oncogene. 18 (48): 6667–76. doi:10.1038/sj.onc.1203120. PMID 10597272. S2CID 25419130.
- Chen S, Nagy PL, Zalkin H (May 1997). "Role of NRF-1 in bidirectional transcription of the human GPAT-AIRC purine biosynthesis locus". Nucleic Acids Research. 25 (9): 1809–16. doi:10.1093/nar/25.9.1809. PMC 146651. PMID 9108165.
- Stanley W, Chu EH (1978). "Assignment of the gene for phosphoribosylpyrophosphate amidotransferase to the pter leads to q21 region of human chromosome 4". Cytogenetics and Cell Genetics. 22 (1–6): 228–31. doi:10.1159/000130943. PMID 752480.
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Bera AK, Chen S, Smith JL, Zalkin H (Dec 1999). "Interdomain signaling in glutamine phosphoribosylpyrophosphate amidotransferase". The Journal of Biological Chemistry. 274 (51): 36498–504. doi:10.1074/jbc.274.51.36498. PMID 10593947. S2CID 46014989.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Zalkin H, Dixon JE (1992). "De novo purine nucleotide biosynthesis". Progress in Nucleic Acid Research and Molecular Biology. 42: 259–87. doi:10.1016/s0079-6603(08)60578-4. ISBN 9780125400428. PMID 1574589.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
External links
[edit]- Amidophosphoribosyl transferase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.