Local adaptation
Local adaptation is a mechanism in evolutionary biology whereby a population of organisms evolves to be more well-suited to its local environment than other members of the same species that live elsewhere. Local adaptation requires that different populations of the same species experience different natural selection. For example, if a species lives across a wide range of temperatures, populations from warm areas may have better heat tolerance than populations of the same species that live in the cold part of its geographic range.
Definition
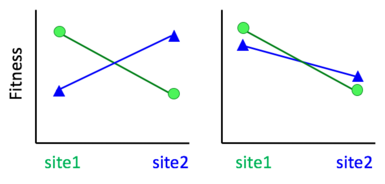
[edit]More formally, a population is said to be locally adapted[1] if organisms in that population have evolved different phenotypes than other populations of the same species, and local phenotypes have higher fitness in their home environment compared to individuals that originate from other locations in the species range.[2][3] This is sometimes called 'home site advantage'.[4] A stricter definition of local adaptation requires 'reciprocal home site advantage', where for a pair of populations each out performs the other in its home site.[5][2] This definition requires that local adaptation result in a fitness trade-off, such that adapting to one environment comes at the cost of poorer performance in a different environment.[3] Before 2004, reciprocal transplants sometimes considered populations locally adapted if the population experienced its highest fitness in its home site vs the foreign site (i.e. compared the same population at multiple sites, vs. multiple populations at the same site). This definition of local adaptation has been largely abandoned after Kawecki and Ebert argued convincingly that populations could be adapted to poor-quality sites but still experience higher fitness if moved to a more benign site (right panel of figure).[3]
Testing for local adaptation
[edit]Testing for local adaptation requires measuring the fitness of organisms from one population in both their local environment and in foreign environments. This is often done using transplant experiments. Using the stricter definition of reciprocal home site advantage, local adaptation is often tested via reciprocal transplant experiments. In reciprocal transplants, organisms from one population are transplanted into another population, and vice versa, and their fitness is measured (see figure).[3] If local transplants outperform (i.e. have higher fitness than) the foreign transplants at both sites, the local populations are said to be locally adapted.[3] If local adaptation is defined simply as a home site advantage of one population (local sources outperform foreign sources at a common site), it can be tested for using common garden experiments, where multiple source populations are grown in a common site, as long as one of the source populations is local to that site.
Transplant experiments have most often been done with plants or other organisms that do not move.[5] However, evidence for rapid local adaptation in mobile animals has been gathered through transplant experiments with Trinidadian guppies.[6]
Frequency of local adaptation
[edit]Several meta-analyses have attempted to quantify how common local adaptation is, and generally reach similar conclusions. Roughly 75% of transplant experiments (mostly with plants) find that local populations outcompete foreign populations at a common site, but less than 50% find the reciprocal home site advantage that defines classic local adaptation.[5][7] Exotic plants are locally adapted to their invasive range as often and as strongly as native plant are locally adapted, suggesting that local adaptation can evolve relatively rapidly.[8][9] However, biologists likely test for local adaptation where they expect to find it. Thus these numbers likely reflect local adaptation between obviously differing sites, rather than the probability than any two randomly-selected populations within a species are locally adapted.
Drivers of local adaptation
[edit]Any component of the environment can drive local adaptation, as long as it affects fitness differently at different sites (creating divergent selection among sites), and does so consistently enough for populations to evolve in response. Seminal examples of local adaptation come from plants that adapted to different elevations[10] or to tolerate heavy metals in soils.[11] Interactions among species (e.g. herbivore-plant interactions) can also drive local adaptation, though do not seem to be as important as abiotic factors, at least for plants in temperate ecosystems.[12] Many examples of local adaptation exist in host-parasite systems as well. For instance, a host may be resistant to a locally-abundant pathogen or parasite, but conspecific hosts from elsewhere where that pathogen is not abundant may have no evolved no such adaptation.[13]
Effects of Gene Flow on Local Adaptation
[edit]Gene flow can completely prevent local adaptations in populations by increasing the amount of genetic material exchanged which can than lower the frequency of alleles associated with the specific local adaptation.[14] However gene flow can also introduce beneficial alleles to a population, which increases the amount of genetic variation, therefore strengthening the likelihood of local adaptations.[15] Gene flow is the transfer of genetic information from one population to another, mainly through migration of organisms or their genetic material.[16] It is possible for genetic material such as pollen or spores that can travel via wind, water or being brought by an animal, to reach an isolated population.[15]
The role gene flow plays in local adaptation is complex because gene flow can reduce the likelihood of local adaptation in a population since gene flow is genetic material from different populations mixing frequently, which makes populations genetically more similar which is the opposite of local adaptation.[17] The level of gene flow impacts its effects on local adaptation, high gene flow tends to reduce local adaptation whereas low gene flow can increase local adaptation.[17] High gene flow is when there is a lot of new genetic material entering the population often and low gene flow is when a population occasionally gets new genetic material. Populations with extensive local adaptations are the most impacted by high gene flow; in such cases where high gene flow occurs in populations with local adaptations it has negative effects such as reducing or removing the adaptation.[14]
Populations with local adaptation can be isolated from other populations however complete isolation is not necessary, gene flow can play a role in populations developing local adaptations. Gene flow allows for the introduction of new beneficial alleles into populations where it was not previously present; if these end up being extremely beneficial to the population they were introduced to, this may allow organisms to locally adapt.[14] Further, local adaptation can happen under gene flow if recombination at genes connected to or controlling the adapted trait is reduced.[14]
Field Observations
[edit]Paper Wasps: Parasite & Host Relationship
[edit]The effect of high gene flow on local adaptation in populations co-evolving with a parasite is of particular interest because parasites are known to specialize on a given host.[14] Populations of coevolving wasps were studied, a type of paper wasps (Polistes biglumis) and the parasite wasp (Polistes atrimandibularis) that preys on it, the parasite essentially takes over the nest of the host and begins to reproduce, eventually taking over the host’s nest.[14] The specific type of parasitism taking place between these two wasp species is social parasitism, meaning one species gets another species to raise its young; social parasitism is known to impact genetic diversity of the host populations.[18] A specific local adaptation of the P. biglumis is having a small number of offspring and putting more energy towards defenses against potential intruders, which would help prevent the parasitic wasp from entering the nest.[14]
Looking at different local populations with similar levels of gene flow is particularly important because the presence of local adaptations in some populations but not others could suggest factors other than gene flow and selective pressure from parasites are causing the differences. Further, regional populations with varying levels of gene flow allows us to get a better idea of how gene flow at the local population level within these regions contributes to local adaptations at the regional level. The Alps were chosen as the area for the wasp study because the elevation of the mountains separate regional and local populations; resulting in multiple local populations of both host and parasite at different elevations and regions.[14] For example, wasps on the same mountain but at different elevations do interbred so gene flow is occurring between local populations. In addition, there are also more isolated regional populations of both host wasp and parasitic wasp on completely different mountains that do not interbreed with other regional populations.[14] DNA microsatellites, a type of genetic marker, were used to study the differences between local populations, to compare to regional populations, in an attempt to see how gene flow was impacting their genetics.[14] What's very important to note is that gene flow is taking place between wasp populations to the same degree; all local populations in the same region have the same amount of gene flow.[14] Meaning that one host population does not have more exposure to different additional genetic material than another host population at a different elevation.
The wasp study found that significant local adaptation only took place in different regional populations, rather than different local populations, for instance higher and lower elevation populations on the same side of the mountain did not have significant differences.[14] But populations in different regions, on the other side of the mountain, a completely different mountain, did have significant differences.[14] Results from the DNA microsatellites showed that the out of the regional wasp populations, the most isolated regional population was the most different from other regional populations.[14] This evidence supports the idea that some level of isolation is needed in order for local adaptations to occur within populations, further supporting the idea that high levels of gene flow do not produce local adaptations.
Experimental Evidence
[edit]Fruit Flies
[edit]Experimental data suggests limited gene flow will produce the most local adaptations and high gene flow will cause populations to hybridize. There was study done on fruit flies (Drosophila melanogaster) to see if adaptive potential was increased in populations that were previously isolated and then experienced different levels of gene flow, or complete hybridization between two populations of previously isolated fruit flies.[17] Experiments introducing different levels of gene flow and complete hydration of D. melanogaster populations showed that limited gene flow (in comparison to high gene flow or full hybridization) was actually what produced the greatest number of beneficial alleles within the fruit fly population.[17]
See also
[edit]References
[edit]- ^ Williams, George (1966). Adaptation and Natural Selection. Princeton: Princeton University Press.
- ^ a b Leimu, Roosa (December 23, 2008). "A meta-analysis of local adaptation in plants". PLOS ONE. 3 (12): e4010. Bibcode:2008PLoSO...3.4010L. doi:10.1371/journal.pone.0004010. PMC 2602971. PMID 19104660.
- ^ a b c d e Kawecki, Tadeusz J.; Ebert, Dieter (2004-12-01). "Conceptual issues in local adaptation" (PDF). Ecology Letters. 7 (12): 1225–1241. Bibcode:2004EcolL...7.1225K. doi:10.1111/j.1461-0248.2004.00684.x. ISSN 1461-0248.
- ^ Galloway, Laura F.; Fenster, Charles B. (2000). "Population Differentiation in an Annual Legume: Local Adaptation". Evolution. 54 (4): 1173–1181. doi:10.1111/j.0014-3820.2000.tb00552.x. ISSN 1558-5646. PMID 11005286. S2CID 13652390.
- ^ a b c Hereford, Joe (2009). "A Quantitative Survey of Local Adaptation and Fitness Trade-Offs". The American Naturalist. 173 (5): 579–588. Bibcode:2009ANat..173..579H. doi:10.1086/597611. ISSN 0003-0147. PMID 19272016. S2CID 524423.
- ^ Gordon, Swanne; Reznick, David; Kinnison, Michael; Bryant, Michael; Weese, Dylan; Räsänen, Katja; Millar, Nathan; Hendry, Andrew (2009). "Adaptive changes in life history and survival following a new guppy introduction". The American Naturalist. 174 (1): 34–45. Bibcode:2009ANat..174...34G. doi:10.1086/599300. PMID 19438322. S2CID 8589987.
- ^ Leimu, Roosa; Fischer, Markus (2008). Buckling, Angus (ed.). "A Meta-Analysis of Local Adaptation in Plants". PLOS ONE. 3 (12): e4010. Bibcode:2008PLoSO...3.4010L. doi:10.1371/journal.pone.0004010. ISSN 1932-6203. PMC 2602971. PMID 19104660.
- ^ Oduor, Ayub M. O.; Leimu, Roosa; Kleunen, Mark van (2016). "Invasive plant species are locally adapted just as frequently and at least as strongly as native plant species". Journal of Ecology. 104 (4): 957–968. Bibcode:2016JEcol.104..957O. doi:10.1111/1365-2745.12578. ISSN 1365-2745.
- ^ Elizabeth, Leger (2009). "Genetic variation and local adaptation at a cheatgrass (Bromus tectorum) invasion edge in western Nevada". Molecular Ecology. 18 (21): 4366–4379. Bibcode:2009MolEc..18.4366L. doi:10.1111/j.1365-294x.2009.04357.x. PMID 19769691. S2CID 13846376.
- ^ Clausen, J (1949). "Experimental Studies on the Nature of Species III. Environmental Responses of Climatic Races of Achillea. Jens Clausen, David D. Keck, William M. Hiesey". The Quarterly Review of Biology. 24 (2): 144. doi:10.1086/396966. ISSN 0033-5770.
- ^ Wilcox Wright, Jessica; Stanton M; Scherson R (2006). "Local adaptation to serpentine and non-serpentine soils in Collinsia sparsiflora". Evolutionary Ecology Research. 8: 1–21.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hargreaves, Anna L.; Germain, Rachel M.; Bontrager, Megan; Persi, Joshua; Angert, Amy L. (2020-03-01). "Local Adaptation to Biotic Interactions: A Meta-analysis across Latitudes". The American Naturalist. 195 (3): 395–411. Bibcode:2020ANat..195..395H. doi:10.1086/707323. ISSN 0003-0147. PMID 32097037. S2CID 209575982.
- ^ Kaltz, O; Shykoff, JA (1998). "Local adaptation in host-parasite systems". Heredity. 81 (4): 361–370. doi:10.1046/j.1365-2540.1998.00435.x.
- ^ a b c d e f g h i j k l m n Seppä, Perttu; Bonelli, Mariaelena; Dupont, Simon; Hakala, Sanja Maria; Bagnères, Anne-Geneviève; Lorenzi, Maria Cristina (September 2020). "Strong Gene Flow Undermines Local Adaptations in a Host Parasite System". Insects. 11 (9): 585. doi:10.3390/insects11090585. ISSN 2075-4450. PMC 7564341. PMID 32882832.
- ^ a b Kling, Matthew M.; Ackerly, David D. (2021-04-27). "Global wind patterns shape genetic differentiation, asymmetric gene flow, and genetic diversity in trees". Proceedings of the National Academy of Sciences. 118 (17). Bibcode:2021PNAS..11817317K. doi:10.1073/pnas.2017317118. ISSN 0027-8424. PMC 8092467. PMID 33875589.
- ^ Choudhuri, Supratim (2014-01-01), Choudhuri, Supratim (ed.), "Chapter 2 - Fundamentals of Molecular Evolution**The opinions expressed in this chapter are the author's own and they do not necessarily reflect the opinions of the FDA, the DHHS, or the Federal Government.", Bioinformatics for Beginners, Oxford: Academic Press, pp. 27–53, doi:10.1016/b978-0-12-410471-6.00002-5, ISBN 978-0-12-410471-6, retrieved 2023-12-11
- ^ a b c d Swindell, William R.; Bouzat, Juan L. (2006-02-01). "Gene Flow and Adaptive Potential in Drosophila melanogaster". Conservation Genetics. 7 (1): 79–89. Bibcode:2006ConG....7...79S. doi:10.1007/s10592-005-8223-5. ISSN 1572-9737. S2CID 25145342.
- ^ Hoffman, Eric A.; Kovacs, Jennifer L.; Goodisman, Michael AD (2008-08-20). "Genetic structure and breeding system in a social wasp and its social parasite". BMC Evolutionary Biology. 8 (1): 239. Bibcode:2008BMCEE...8..239H. doi:10.1186/1471-2148-8-239. ISSN 1471-2148. PMC 2533669. PMID 18715511.
