Mitochondrion
| Mitochondrion | |
|---|---|
 Diagram of animal mitochondrion | |
| Details | |
| Pronunciation | /ˌmaɪtəˈkɒndriən/[1] |
| Part of | Cell |
| Identifiers | |
| Latin | organella |
| MeSH | D008928 |
| FMA | 63835 |
| Anatomical terms of microanatomy | |
| Cell biology | |
|---|---|
| Animal cell diagram | |
 Components of a typical animal cell:
|
A mitochondrion (pl. mitochondria) is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy.[2] They were discovered by Albert von Kölliker in 1857[3] in the voluntary muscles of insects. Meaning a thread-like granule, the term mitochondrion was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 Scientific American article of the same name.[4]
Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal Henneguya salminicola is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome.[5][6][7] A large number of unicellular organisms, such as microsporidia, parabasalids and diplomonads, have reduced or transformed their mitochondria into other structures,[8] e.g. hydrogenosomes and mitosomes.[9] The oxymonads Monocercomonoides, Streblomastix, and Blattamonas have completely lost their mitochondria.[5][10]
Mitochondria are commonly between 0.75 and 3 μm2 in cross section,[11] but vary considerably in size and structure. Unless specifically stained, they are not visible. In addition to supplying cellular energy, mitochondria are involved in other tasks, such as signaling, cellular differentiation, and cell death, as well as maintaining control of the cell cycle and cell growth.[12] Mitochondrial biogenesis is in turn temporally coordinated with these cellular processes.[13][14] Mitochondria have been implicated in several human disorders and conditions, such as mitochondrial diseases,[15] cardiac dysfunction,[16] heart failure[17] and autism.[18]
The number of mitochondria in a cell can vary widely by organism, tissue, and cell type. A mature red blood cell has no mitochondria,[19] whereas a liver cell can have more than 2000.[20][21] The mitochondrion is composed of compartments that carry out specialized functions. These compartments or regions include the outer membrane, intermembrane space, inner membrane, cristae, and matrix.
Although most of a eukaryotic cell's DNA is contained in the cell nucleus, the mitochondrion has its own genome ("mitogenome") that is substantially similar to bacterial genomes.[22] This finding has led to general acceptance of the endosymbiotic hypothesis - that free-living prokaryotic ancestors of modern mitochondria permanently fused with eukaryotic cells in the distant past, evolving such that modern animals, plants, fungi, and other eukaryotes are able to respire to generate cellular energy.[23]
Structure
Mitochondria may have a number of different shapes.[24] A mitochondrion contains outer and inner membranes composed of phospholipid bilayers and proteins.[20] The two membranes have different properties. Because of this double-membraned organization, there are five distinct parts to a mitochondrion:
- The outer mitochondrial membrane,
- The intermembrane space (the space between the outer and inner membranes),
- The inner mitochondrial membrane,
- The cristae space (formed by infoldings of the inner membrane), and
- The matrix (space within the inner membrane), which is a fluid.
Mitochondria have folding to increase surface area, which in turn increases ATP (adenosine triphosphate) production. Mitochondria stripped of their outer membrane are called mitoplasts.
Outer membrane
The outer mitochondrial membrane, which encloses the entire organelle, is 60 to 75 angstroms (Å) thick. It has a protein-to-phospholipid ratio similar to that of the cell membrane (about 1:1 by weight). It contains large numbers of integral membrane proteins called porins. A major trafficking protein is the pore-forming voltage-dependent anion channel (VDAC). The VDAC is the primary transporter of nucleotides, ions and metabolites between the cytosol and the intermembrane space.[25][26] It is formed as a beta barrel that spans the outer membrane, similar to that in the gram-negative bacterial outer membrane.[27] Larger proteins can enter the mitochondrion if a signaling sequence at their N-terminus binds to a large multisubunit protein called translocase in the outer membrane, which then actively moves them across the membrane.[28] Mitochondrial pro-proteins are imported through specialised translocation complexes.
The outer membrane also contains enzymes involved in such diverse activities as the elongation of fatty acids, oxidation of epinephrine, and the degradation of tryptophan. These enzymes include monoamine oxidase, rotenone-insensitive NADH-cytochrome c-reductase, kynurenine hydroxylase and fatty acid Co-A ligase. Disruption of the outer membrane permits proteins in the intermembrane space to leak into the cytosol, leading to cell death.[29] The outer mitochondrial membrane can associate with the endoplasmic reticulum (ER) membrane, in a structure called MAM (mitochondria-associated ER-membrane). This is important in the ER-mitochondria calcium signaling and is involved in the transfer of lipids between the ER and mitochondria.[30] Outside the outer membrane are small (diameter: 60 Å) particles named sub-units of Parson.
Intermembrane space
The mitochondrial intermembrane space is the space between the outer membrane and the inner membrane. It is also known as perimitochondrial space. Because the outer membrane is freely permeable to small molecules, the concentrations of small molecules, such as ions and sugars, in the intermembrane space is the same as in the cytosol.[20] However, large proteins must have a specific signaling sequence to be transported across the outer membrane, so the protein composition of this space is different from the protein composition of the cytosol. One protein that is localized to the intermembrane space in this way is cytochrome c.[29]
Inner membrane
The inner mitochondrial membrane contains proteins with three types of functions:[20]
- Those that perform the electron transport chain redox reactions
- ATP synthase, which generates ATP in the matrix
- Specific transport proteins that regulate metabolite passage into and out of the mitochondrial matrix
It contains more than 151 different polypeptides, and has a very high protein-to-phospholipid ratio (more than 3:1 by weight, which is about 1 protein for 15 phospholipids). The inner membrane is home to around 1/5 of the total protein in a mitochondrion.[31] Additionally, the inner membrane is rich in an unusual phospholipid, cardiolipin. This phospholipid was originally discovered in cow hearts in 1942, and is usually characteristic of mitochondrial and bacterial plasma membranes.[32] Cardiolipin contains four fatty acids rather than two, and may help to make the inner membrane impermeable,[20] and its disruption can lead to multiple clinical disorders including neurological disorders and cancer.[33] Unlike the outer membrane, the inner membrane does not contain porins, and is highly impermeable to all molecules. Almost all ions and molecules require special membrane transporters to enter or exit the matrix. Proteins are ferried into the matrix via the translocase of the inner membrane (TIM) complex or via OXA1L.[28] In addition, there is a membrane potential across the inner membrane, formed by the action of the enzymes of the electron transport chain. Inner membrane fusion is mediated by the inner membrane protein OPA1.[34]
Cristae
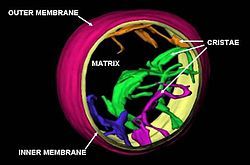
The inner mitochondrial membrane is compartmentalized into numerous folds called cristae, which expand the surface area of the inner mitochondrial membrane, enhancing its ability to produce ATP. For typical liver mitochondria, the area of the inner membrane is about five times as large as that of the outer membrane. This ratio is variable and mitochondria from cells that have a greater demand for ATP, such as muscle cells, contain even more cristae. Mitochondria within the same cell can have substantially different crista-density, with the ones that are required to produce more energy having much more crista-membrane surface.[35] These folds are studded with small round bodies known as F1 particles or oxysomes.[36]
Matrix
The matrix is the space enclosed by the inner membrane. It contains about 2/3 of the total proteins in a mitochondrion.[20] The matrix is important in the production of ATP with the aid of the ATP synthase contained in the inner membrane. The matrix contains a highly concentrated mixture of hundreds of enzymes, special mitochondrial ribosomes, tRNA, and several copies of the mitochondrial DNA genome. Of the enzymes, the major functions include oxidation of pyruvate and fatty acids, and the citric acid cycle.[20] The DNA molecules are packaged into nucleoids by proteins, one of which is TFAM.[37]
Function
The most prominent roles of mitochondria are to produce the energy currency of the cell, ATP (i.e., phosphorylation of ADP), through respiration and to regulate cellular metabolism.[21] The central set of reactions involved in ATP production are collectively known as the citric acid cycle, or the Krebs cycle, and oxidative phosphorylation. However, the mitochondrion has many other functions in addition to the production of ATP.
Energy conversion
A dominant role for the mitochondria is the production of ATP, as reflected by the large number of proteins in the inner membrane for this task. This is done by oxidizing the major products of glucose: pyruvate, and NADH, which are produced in the cytosol.[21] This type of cellular respiration, known as aerobic respiration, is dependent on the presence of oxygen. When oxygen is limited, the glycolytic products will be metabolized by anaerobic fermentation, a process that is independent of the mitochondria.[21] The production of ATP from glucose and oxygen has an approximately 13-times higher yield during aerobic respiration compared to fermentation.[38] Plant mitochondria can also produce a limited amount of ATP either by breaking the sugar produced during photosynthesis or without oxygen by using the alternate substrate nitrite.[39] ATP crosses out through the inner membrane with the help of a specific protein, and across the outer membrane via porins.[40] After conversion of ATP to ADP by dephosphorylation that releases energy, ADP returns via the same route.
Pyruvate and the citric acid cycle
Pyruvate molecules produced by glycolysis are actively transported across the inner mitochondrial membrane, and into the matrix where they can either be oxidized and combined with coenzyme A to form CO2, acetyl-CoA, and NADH,[21] or they can be carboxylated (by pyruvate carboxylase) to form oxaloacetate. This latter reaction "fills up" the amount of oxaloacetate in the citric acid cycle and is therefore an anaplerotic reaction, increasing the cycle's capacity to metabolize acetyl-CoA when the tissue's energy needs (e.g., in muscle) are suddenly increased by activity.[41]
In the citric acid cycle, all the intermediates (e.g. citrate, iso-citrate, alpha-ketoglutarate, succinate, fumarate, malate and oxaloacetate) are regenerated during each turn of the cycle. Adding more of any of these intermediates to the mitochondrion therefore means that the additional amount is retained within the cycle, increasing all the other intermediates as one is converted into the other. Hence, the addition of any one of them to the cycle has an anaplerotic effect, and its removal has a cataplerotic effect. These anaplerotic and cataplerotic reactions will, during the course of the cycle, increase or decrease the amount of oxaloacetate available to combine with acetyl-CoA to form citric acid. This in turn increases or decreases the rate of ATP production by the mitochondrion, and thus the availability of ATP to the cell.[41]
Acetyl-CoA, on the other hand, derived from pyruvate oxidation, or from the beta-oxidation of fatty acids, is the only fuel to enter the citric acid cycle. With each turn of the cycle one molecule of acetyl-CoA is consumed for every molecule of oxaloacetate present in the mitochondrial matrix, and is never regenerated. It is the oxidation of the acetate portion of acetyl-CoA that produces CO2 and water, with the energy thus released captured in the form of ATP.[41]
In the liver, the carboxylation of cytosolic pyruvate into intra-mitochondrial oxaloacetate is an early step in the gluconeogenic pathway, which converts lactate and de-aminated alanine into glucose,[21][41] under the influence of high levels of glucagon and/or epinephrine in the blood.[41] Here, the addition of oxaloacetate to the mitochondrion does not have a net anaplerotic effect, as another citric acid cycle intermediate (malate) is immediately removed from the mitochondrion to be converted to cytosolic oxaloacetate, and ultimately to glucose, in a process that is almost the reverse of glycolysis.[41]
The enzymes of the citric acid cycle are located in the mitochondrial matrix, with the exception of succinate dehydrogenase, which is bound to the inner mitochondrial membrane as part of Complex II.[42] The citric acid cycle oxidizes the acetyl-CoA to carbon dioxide, and, in the process, produces reduced cofactors (three molecules of NADH and one molecule of FADH2) that are a source of electrons for the electron transport chain, and a molecule of GTP (which is readily converted to an ATP).[21]
O2 and NADH: energy-releasing reactions

The electrons from NADH and FADH2 are transferred to oxygen (O2) and hydrogen (protons) in several steps via an electron transport chain. NADH and FADH2 molecules are produced within the matrix via the citric acid cycle and in the cytoplasm by glycolysis. Reducing equivalents from the cytoplasm can be imported via the malate-aspartate shuttle system of antiporter proteins or fed into the electron transport chain using a glycerol phosphate shuttle.[21]
The major energy-releasing reactions[43][44] that make the mitochondrion the "powerhouse of the cell" occur at protein complexes I, III and IV in the inner mitochondrial membrane (NADH dehydrogenase (ubiquinone), cytochrome c reductase, and cytochrome c oxidase). At complex IV, O2 reacts with the reduced form of iron in cytochrome c:
releasing a lot of free energy[44][43] from the reactants without breaking bonds of an organic fuel. The free energy put in to remove an electron from Fe2+ is released at complex III when Fe3+ of cytochrome c reacts to oxidize ubiquinol (QH2):
The ubiquinone (Q) generated reacts, in complex I, with NADH:
While the reactions are controlled by an electron transport chain, free electrons are not amongst the reactants or products in the three reactions shown and therefore do not affect the free energy released, which is used to pump protons (H+) into the intermembrane space. This process is efficient, but a small percentage of electrons may prematurely reduce oxygen, forming reactive oxygen species such as superoxide.[21] This can cause oxidative stress in the mitochondria and may contribute to the decline in mitochondrial function associated with aging.[45]
As the proton concentration increases in the intermembrane space, a strong electrochemical gradient is established across the inner membrane. The protons can return to the matrix through the ATP synthase complex, and their potential energy is used to synthesize ATP from ADP and inorganic phosphate (Pi).[21] This process is called chemiosmosis, and was first described by Peter Mitchell,[46][47] who was awarded the 1978 Nobel Prize in Chemistry for his work. Later, part of the 1997 Nobel Prize in Chemistry was awarded to Paul D. Boyer and John E. Walker for their clarification of the working mechanism of ATP synthase.[48]
Heat production
Under certain conditions, protons can re-enter the mitochondrial matrix without contributing to ATP synthesis. This process is known as proton leak or mitochondrial uncoupling and is due to the facilitated diffusion of protons into the matrix. The process results in the unharnessed potential energy of the proton electrochemical gradient being released as heat.[21] The process is mediated by a proton channel called thermogenin, or UCP1.[49] Thermogenin is primarily found in brown adipose tissue, or brown fat, and is responsible for non-shivering thermogenesis. Brown adipose tissue is found in mammals, and is at its highest levels in early life and in hibernating animals. In humans, brown adipose tissue is present at birth and decreases with age.[49]
Mitochondrial fatty acid synthesis
Mitochondrial fatty acid synthesis (mtFASII) is essential for cellular respiration and mitochondrial biogenesis.[50] It is also thought to play a role as a mediator in intracellular signaling due to its influence on the levels of bioactive lipids, such as lysophospholipids and sphingolipids.[51]
Octanoyl-ACP (C8) is considered to be the most important end product of mtFASII, which also forms the starting substrate of lipoic acid biosynthesis.[52] Since lipoic acid is the cofactor of important mitochondrial enzyme complexes, such as the pyruvate dehydrogenase complex (PDC), α-ketoglutarate dehydrogenase complex (OGDC), branched-chain α-ketoacid dehydrogenase complex (BCKDC), and in the glycine cleavage system (GCS), mtFASII has an influence on energy metabolism.[53]
Other products of mtFASII play a role in the regulation of mitochondrial translation, FeS cluster biogenesis and assembly of oxidative phosphorylation complexes.[52]
Furthermore, with the help of mtFASII and acylated ACP, acetyl-CoA regulates its consumption in mitochondria.[52]
Uptake, storage and release of calcium ions
The concentrations of free calcium in the cell can regulate an array of reactions and is important for signal transduction in the cell. Mitochondria can transiently store calcium, a contributing process for the cell's homeostasis of calcium.[54] [55] Their ability to rapidly take in calcium for later release makes them good "cytosolic buffers" for calcium.[56][57][58] The endoplasmic reticulum (ER) is the most significant storage site of calcium,[59] and there is a significant interplay between the mitochondrion and ER with regard to calcium.[60] The calcium is taken up into the matrix by the mitochondrial calcium uniporter on the inner mitochondrial membrane.[61] It is primarily driven by the mitochondrial membrane potential.[55] Release of this calcium back into the cell's interior can occur via a sodium-calcium exchange protein or via "calcium-induced-calcium-release" pathways.[61] This can initiate calcium spikes or calcium waves with large changes in the membrane potential. These can activate a series of second messenger system proteins that can coordinate processes such as neurotransmitter release in nerve cells and release of hormones in endocrine cells.[62]
Ca2+ influx to the mitochondrial matrix has recently been implicated as a mechanism to regulate respiratory bioenergetics by allowing the electrochemical potential across the membrane to transiently "pulse" from ΔΨ-dominated to pH-dominated, facilitating a reduction of oxidative stress.[63] In neurons, concomitant increases in cytosolic and mitochondrial calcium act to synchronize neuronal activity with mitochondrial energy metabolism. Mitochondrial matrix calcium levels can reach the tens of micromolar levels, which is necessary for the activation of isocitrate dehydrogenase, one of the key regulatory enzymes of the Krebs cycle.[64]
Cellular proliferation regulation
The relationship between cellular proliferation and mitochondria has been investigated. Tumor cells require ample ATP to synthesize bioactive compounds such as lipids, proteins, and nucleotides for rapid proliferation.[65] The majority of ATP in tumor cells is generated via the oxidative phosphorylation pathway (OxPhos).[66] Interference with OxPhos cause cell cycle arrest suggesting that mitochondria play a role in cell proliferation.[66] Mitochondrial ATP production is also vital for cell division and differentiation in infection[67] in addition to basic functions in the cell including the regulation of cell volume, solute concentration, and cellular architecture.[68][69][70] ATP levels differ at various stages of the cell cycle suggesting that there is a relationship between the abundance of ATP and the cell's ability to enter a new cell cycle.[71] ATP's role in the basic functions of the cell make the cell cycle sensitive to changes in the availability of mitochondrial derived ATP.[71] The variation in ATP levels at different stages of the cell cycle support the hypothesis that mitochondria play an important role in cell cycle regulation.[71] Although the specific mechanisms between mitochondria and the cell cycle regulation is not well understood, studies have shown that low energy cell cycle checkpoints monitor the energy capability before committing to another round of cell division.[12]
Programmed cell death and innate immunity
Programmed cell death (PCD) is crucial for various physiological functions, including organ development and cellular homeostasis. It serves as an intrinsic mechanism to prevent malignant transformation and plays a fundamental role in immunity by aiding in antiviral defense, pathogen elimination, inflammation, and immune cell recruitment.[72]
Mitochondria have long been recognized for their central role in the intrinsic pathway of apoptosis, a form of PCD.[73] In recent decades, they have also been identified as a signalling hub for much of the innate immune system.[74] The endosymbiotic origin of mitochondria distinguishes them from other cellular components, and the exposure of mitochondrial elements to the cytosol can trigger the same pathways as infection markers. These pathways lead to apoptosis, autophagy, or the induction of proinflammatory genes.[75][74]
Mitochondria contribute to apoptosis by releasing cytochrome c, which directly induces the formation of apoptosomes. Additionally, they are a source of various damage-associated molecular patterns (DAMPs). These DAMPs are often recognised by the same pattern-recognition receptors (PRRs) that respond to pathogen-associated molecular patterns (PAMPs) during infections.[76] For example, mitochondrial mtDNA resembles bacterial DNA due to its lack of CpG methylation and can be detected by Toll-like receptor 9 and cGAS.[77] Double-stranded RNA (dsRNA), produced due to bidirectional mitochondrial transcription, can activate viral sensing pathways through RIG-I-like receptors.[78] Additionally, the N-formylation of mitochondrial proteins, similar to that of bacterial proteins, can be recognized by formyl peptide receptors.[79][80]
Normally, these mitochondrial components are sequestered from the rest of the cell but are released following mitochondrial membrane permeabilization during apoptosis or passively after mitochondrial damage. However, mitochondria also play an active role in innate immunity, releasing mtDNA in response to metabolic cues.[74] Mitochondria are also the localization site for immune and apoptosis regulatory proteins, such as BAX, MAVS (located on the outer membrane), and NLRX1 (found in the matrix). These proteins are modulated by the mitochondrial metabolic status and mitochondrial dynamics.[74][81][82]
Additional functions
Mitochondria play a central role in many other metabolic tasks, such as:
- Signaling through mitochondrial reactive oxygen species[83]
- Regulation of the membrane potential[21]
- Calcium signaling (including calcium-evoked apoptosis)[84]
- Regulation of cellular metabolism[12]
- Certain heme synthesis reactions[85] (see also: Porphyrin)
- Steroid synthesis[56]
- Hormonal signaling[86] – mitochondria are sensitive and responsive to hormones, in part by the action of mitochondrial estrogen receptors (mtERs). These receptors have been found in various tissues and cell types, including brain[87] and heart[88]
- Development and function of immune cells[89]
- Neuronal mitochondria also contribute to cellular quality control by reporting neuronal status towards microglia through specialised somatic-junctions.[90]
- Mitochondria of developing neurons contribute to intercellular signaling towards microglia, which communication is indispensable for proper regulation of brain development.[91]
Some mitochondrial functions are performed only in specific types of cells. For example, mitochondria in liver cells contain enzymes that allow them to detoxify ammonia, a waste product of protein metabolism. A mutation in the genes regulating any of these functions can result in mitochondrial diseases.
Mitochondrial proteins (proteins transcribed from mitochondrial DNA) vary depending on the tissue and the species. In humans, 615 distinct types of proteins have been identified from cardiac mitochondria,[92] whereas in rats, 940 proteins have been reported.[93] The mitochondrial proteome is thought to be dynamically regulated.[94]
Organization and distribution

Mitochondria (or related structures) are found in all eukaryotes (except the Oxymonad Monocercomonoides).[5] Although commonly depicted as bean-like structures they form a highly dynamic network in the majority of cells where they constantly undergo fission and fusion. The population of all the mitochondria of a given cell constitutes the chondriome.[95] Mitochondria vary in number and location according to cell type. A single mitochondrion is often found in unicellular organisms, while human liver cells have about 1000–2000 mitochondria per cell, making up 1/5 of the cell volume.[20] The mitochondrial content of otherwise similar cells can vary substantially in size and membrane potential,[96] with differences arising from sources including uneven partitioning at cell division, leading to extrinsic differences in ATP levels and downstream cellular processes.[97] The mitochondria can be found nestled between myofibrils of muscle or wrapped around the sperm flagellum.[20] Often, they form a complex 3D branching network inside the cell with the cytoskeleton. The association with the cytoskeleton determines mitochondrial shape, which can affect the function as well:[98] different structures of the mitochondrial network may afford the population a variety of physical, chemical, and signalling advantages or disadvantages.[99] Mitochondria in cells are always distributed along microtubules and the distribution of these organelles is also correlated with the endoplasmic reticulum.[100] Recent evidence suggests that vimentin, one of the components of the cytoskeleton, is also critical to the association with the cytoskeleton.[101]
Mitochondria-associated ER membrane (MAM)
The mitochondria-associated ER membrane (MAM) is another structural element that is increasingly recognized for its critical role in cellular physiology and homeostasis. Once considered a technical snag in cell fractionation techniques, the alleged ER vesicle contaminants that invariably appeared in the mitochondrial fraction have been re-identified as membranous structures derived from the MAM—the interface between mitochondria and the ER.[102] Physical coupling between these two organelles had previously been observed in electron micrographs and has more recently been probed with fluorescence microscopy.[102] Such studies estimate that at the MAM, which may comprise up to 20% of the mitochondrial outer membrane, the ER and mitochondria are separated by a mere 10–25 nm and held together by protein tethering complexes.[102][30][103]
Purified MAM from subcellular fractionation is enriched in enzymes involved in phospholipid exchange, in addition to channels associated with Ca2+ signaling.[102][103] These hints of a prominent role for the MAM in the regulation of cellular lipid stores and signal transduction have been borne out, with significant implications for mitochondrial-associated cellular phenomena, as discussed below. Not only has the MAM provided insight into the mechanistic basis underlying such physiological processes as intrinsic apoptosis and the propagation of calcium signaling, but it also favors a more refined view of the mitochondria. Though often seen as static, isolated 'powerhouses' hijacked for cellular metabolism through an ancient endosymbiotic event, the evolution of the MAM underscores the extent to which mitochondria have been integrated into overall cellular physiology, with intimate physical and functional coupling to the endomembrane system.
Phospholipid transfer
The MAM is enriched in enzymes involved in lipid biosynthesis, such as phosphatidylserine synthase on the ER face and phosphatidylserine decarboxylase on the mitochondrial face.[104][105] Because mitochondria are dynamic organelles constantly undergoing fission and fusion events, they require a constant and well-regulated supply of phospholipids for membrane integrity.[106][107] But mitochondria are not only a destination for the phospholipids they finish synthesis of; rather, this organelle also plays a role in inter-organelle trafficking of the intermediates and products of phospholipid biosynthetic pathways, ceramide and cholesterol metabolism, and glycosphingolipid anabolism.[105][107]
Such trafficking capacity depends on the MAM, which has been shown to facilitate transfer of lipid intermediates between organelles.[104] In contrast to the standard vesicular mechanism of lipid transfer, evidence indicates that the physical proximity of the ER and mitochondrial membranes at the MAM allows for lipid flipping between opposed bilayers.[107] Despite this unusual and seemingly energetically unfavorable mechanism, such transport does not require ATP.[107] Instead, in yeast, it has been shown to be dependent on a multiprotein tethering structure termed the ER-mitochondria encounter structure, or ERMES, although it remains unclear whether this structure directly mediates lipid transfer or is required to keep the membranes in sufficiently close proximity to lower the energy barrier for lipid flipping.[107][108]
The MAM may also be part of the secretory pathway, in addition to its role in intracellular lipid trafficking. In particular, the MAM appears to be an intermediate destination between the rough ER and the Golgi in the pathway that leads to very-low-density lipoprotein, or VLDL, assembly and secretion.[105][109] The MAM thus serves as a critical metabolic and trafficking hub in lipid metabolism.
Calcium signaling
A critical role for the ER in calcium signaling was acknowledged before such a role for the mitochondria was widely accepted, in part because the low affinity of Ca2+ channels localized to the outer mitochondrial membrane seemed to contradict this organelle's purported responsiveness to changes in intracellular Ca2+ flux.[102][59] But the presence of the MAM resolves this apparent contradiction: the close physical association between the two organelles results in Ca2+ microdomains at contact points that facilitate efficient Ca2+ transmission from the ER to the mitochondria.[102] Transmission occurs in response to so-called "Ca2+ puffs" generated by spontaneous clustering and activation of IP3R, a canonical ER membrane Ca2+ channel.[102][30]
The fate of these puffs—in particular, whether they remain restricted to isolated locales or integrated into Ca2+ waves for propagation throughout the cell—is determined in large part by MAM dynamics. Although reuptake of Ca2+ by the ER (concomitant with its release) modulates the intensity of the puffs, thus insulating mitochondria to a certain degree from high Ca2+ exposure, the MAM often serves as a firewall that essentially buffers Ca2+ puffs by acting as a sink into which free ions released into the cytosol can be funneled.[102][110][111] This Ca2+ tunneling occurs through the low-affinity Ca2+ receptor VDAC1, which recently has been shown to be physically tethered to the IP3R clusters on the ER membrane and enriched at the MAM.[102][30][112] The ability of mitochondria to serve as a Ca2+ sink is a result of the electrochemical gradient generated during oxidative phosphorylation, which makes tunneling of the cation an exergonic process.[112] Normal, mild calcium influx from cytosol into the mitochondrial matrix causes transient depolarization that is corrected by pumping out protons.
But transmission of Ca2+ is not unidirectional; rather, it is a two-way street.[59] The properties of the Ca2+ pump SERCA and the channel IP3R present on the ER membrane facilitate feedback regulation coordinated by MAM function. In particular, the clearance of Ca2+ by the MAM allows for spatio-temporal patterning of Ca2+ signaling because Ca2+ alters IP3R activity in a biphasic manner.[102] SERCA is likewise affected by mitochondrial feedback: uptake of Ca2+ by the MAM stimulates ATP production, thus providing energy that enables SERCA to reload the ER with Ca2+ for continued Ca2+ efflux at the MAM.[110][112] Thus, the MAM is not a passive buffer for Ca2+ puffs; rather it helps modulate further Ca2+ signaling through feedback loops that affect ER dynamics.
Regulating ER release of Ca2+ at the MAM is especially critical because only a certain window of Ca2+ uptake sustains the mitochondria, and consequently the cell, at homeostasis. Sufficient intraorganelle Ca2+ signaling is required to stimulate metabolism by activating dehydrogenase enzymes critical to flux through the citric acid cycle.[113][114] However, once Ca2+ signaling in the mitochondria passes a certain threshold, it stimulates the intrinsic pathway of apoptosis in part by collapsing the mitochondrial membrane potential required for metabolism.[102] Studies examining the role of pro- and anti-apoptotic factors support this model; for example, the anti-apoptotic factor Bcl-2 has been shown to interact with IP3Rs to reduce Ca2+ filling of the ER, leading to reduced efflux at the MAM and preventing collapse of the mitochondrial membrane potential post-apoptotic stimuli.[102] Given the need for such fine regulation of Ca2+ signaling, it is perhaps unsurprising that dysregulated mitochondrial Ca2+ has been implicated in several neurodegenerative diseases, while the catalogue of tumor suppressors includes a few that are enriched at the MAM.[112]
Molecular basis for tethering
Recent advances in the identification of the tethers between the mitochondrial and ER membranes suggest that the scaffolding function of the molecular elements involved is secondary to other, non-structural functions. In yeast, ERMES, a multiprotein complex of interacting ER- and mitochondrial-resident membrane proteins, is required for lipid transfer at the MAM and exemplifies this principle. One of its components, for example, is also a constituent of the protein complex required for insertion of transmembrane beta-barrel proteins into the lipid bilayer.[107] However, a homologue of the ERMES complex has not yet been identified in mammalian cells. Other proteins implicated in scaffolding likewise have functions independent of structural tethering at the MAM; for example, ER-resident and mitochondrial-resident mitofusins form heterocomplexes that regulate the number of inter-organelle contact sites, although mitofusins were first identified for their role in fission and fusion events between individual mitochondria.[102] Glucose-related protein 75 (grp75) is another dual-function protein. In addition to the matrix pool of grp75, a portion serves as a chaperone that physically links the mitochondrial and ER Ca2+ channels VDAC and IP3R for efficient Ca2+ transmission at the MAM.[102][30] Another potential tether is Sigma-1R, a non-opioid receptor whose stabilization of ER-resident IP3R may preserve communication at the MAM during the metabolic stress response.[115][116]

Perspective
The MAM is a critical signaling, metabolic, and trafficking hub in the cell that allows for the integration of ER and mitochondrial physiology. Coupling between these organelles is not simply structural but functional as well and critical for overall cellular physiology and homeostasis. The MAM thus offers a perspective on mitochondria that diverges from the traditional view of this organelle as a static, isolated unit appropriated for its metabolic capacity by the cell.[117] Instead, this mitochondrial-ER interface emphasizes the integration of the mitochondria, the product of an endosymbiotic event, into diverse cellular processes. Recently it has also been shown, that mitochondria and MAM-s in neurons are anchored to specialised intercellular communication sites (so called somatic-junctions). Microglial processes monitor and protect neuronal functions at these sites, and MAM-s are supposed to have an important role in this type of cellular quality-control.[90]
Origin and evolution
There are two hypotheses about the origin of mitochondria: endosymbiotic and autogenous. The endosymbiotic hypothesis suggests that mitochondria were originally prokaryotic cells, capable of implementing oxidative mechanisms that were not possible for eukaryotic cells; they became endosymbionts living inside the eukaryote.[23][118][119][120] In the autogenous hypothesis, mitochondria were born by splitting off a portion of DNA from the nucleus of the eukaryotic cell at the time of divergence with the prokaryotes; this DNA portion would have been enclosed by membranes, which could not be crossed by proteins. Since mitochondria have many features in common with bacteria, the endosymbiotic hypothesis is the more widely accepted of the two accounts.[120][121]
A mitochondrion contains DNA, which is organized as several copies of a single, usually circular chromosome. This mitochondrial chromosome contains genes for redox proteins, such as those of the respiratory chain. The CoRR hypothesis proposes that this co-location is required for redox regulation. The mitochondrial genome codes for some RNAs of ribosomes, and the 22 tRNAs necessary for the translation of mRNAs into protein. The circular structure is also found in prokaryotes. The proto-mitochondrion was probably closely related to Rickettsia.[122][123] However, the exact relationship of the ancestor of mitochondria to the alphaproteobacteria and whether the mitochondrion was formed at the same time or after the nucleus, remains controversial.[124] For example, it has been suggested that the SAR11 clade of bacteria shares a relatively recent common ancestor with the mitochondria,[125] while phylogenomic analyses indicate that mitochondria evolved from a Pseudomonadota lineage that is closely related to or a member of alphaproteobacteria.[126][127] Some papers describe mitochondria as sister to the alphaproteobactera, together forming the sister the marineproteo1 group, together forming the sister to Magnetococcidae.[128][129][130][131]
The ribosomes coded for by the mitochondrial DNA are similar to those from bacteria in size and structure.[132] They closely resemble the bacterial 70S ribosome and not the 80S cytoplasmic ribosomes, which are coded for by nuclear DNA.
The endosymbiotic relationship of mitochondria with their host cells was popularized by Lynn Margulis.[133] The endosymbiotic hypothesis suggests that mitochondria descended from aerobic bacteria that somehow survived endocytosis by another cell, and became incorporated into the cytoplasm. The ability of these bacteria to conduct respiration in host cells that had relied on glycolysis and fermentation would have provided a considerable evolutionary advantage. This symbiotic relationship probably developed 1.7 to 2 billion years ago.[134][135]

A few groups of unicellular eukaryotes have only vestigial mitochondria or derived structures: The microsporidians, metamonads, and archamoebae.[136] These groups appear as the most primitive eukaryotes on phylogenetic trees constructed using rRNA information, which once suggested that they appeared before the origin of mitochondria. However, this is now known to be an artifact of long-branch attraction: They are derived groups and retain genes or organelles derived from mitochondria (e. g., mitosomes and hydrogenosomes).[8] Hydrogenosomes, mitosomes, and related organelles as found in some loricifera (e. g. Spinoloricus)[137][138] and myxozoa (e. g. Henneguya zschokkei) are together classified as MROs, mitochondrion-related organelles.[7][139]
Monocercomonoides and other oxymonads appear to have lost their mitochondria completely and at least some of the mitochondrial functions seem to be carried out by cytoplasmic proteins now.[5][140][10]
Mitochondrial genetics

Mitochondria contain their own genome. The human mitochondrial genome is a circular double-stranded DNA molecule of about 16 kilobases.[141] It encodes 37 genes: 13 for subunits of respiratory complexes I, III, IV and V, 22 for mitochondrial tRNA (for the 20 standard amino acids, plus an extra gene for leucine and serine), and 2 for rRNA (12S and 16S rRNA).[141] One mitochondrion can contain two to ten copies of its DNA.[142] One of the two mitochondrial DNA (mtDNA) strands has a disproportionately higher ratio of the heavier nucleotides adenine and guanine, and this is termed the heavy strand (or H strand), whereas the other strand is termed the light strand (or L strand). The weight difference allows the two strands to be separated by centrifugation. mtDNA has one long non-coding stretch known as the non-coding region (NCR), which contains the heavy strand promoter (HSP) and light strand promoter (LSP) for RNA transcription, the origin of replication for the H strand (OriH) localized on the L strand, three conserved sequence boxes (CSBs 1–3), and a termination-associated sequence (TAS). The origin of replication for the L strand (OriL) is localized on the H strand 11,000 bp downstream of OriH, located within a cluster of genes coding for tRNA.[143]
As in prokaryotes, there is a very high proportion of coding DNA and an absence of repeats. Mitochondrial genes are transcribed as multigenic transcripts, which are cleaved and polyadenylated to yield mature mRNAs. Most proteins necessary for mitochondrial function are encoded by genes in the cell nucleus and the corresponding proteins are imported into the mitochondrion.[144] The exact number of genes encoded by the nucleus and the mitochondrial genome differs between species. Most mitochondrial genomes are circular.[145] In general, mitochondrial DNA lacks introns, as is the case in the human mitochondrial genome;[144] however, introns have been observed in some eukaryotic mitochondrial DNA,[146] such as that of yeast[147] and protists,[148] including Dictyostelium discoideum.[149] Between protein-coding regions, tRNAs are present. Mitochondrial tRNA genes have different sequences from the nuclear tRNAs, but lookalikes of mitochondrial tRNAs have been found in the nuclear chromosomes with high sequence similarity.[150]
In animals, the mitochondrial genome is typically a single circular chromosome that is approximately 16 kb long and has 37 genes. The genes, while highly conserved, may vary in location. Curiously, this pattern is not found in the human body louse (Pediculus humanus). Instead, this mitochondrial genome is arranged in 18 minicircular chromosomes, each of which is 3–4 kb long and has one to three genes.[151] This pattern is also found in other sucking lice, but not in chewing lice. Recombination has been shown to occur between the minichromosomes.
Human population genetic studies
The near-absence of genetic recombination in mitochondrial DNA makes it a useful source of information for studying population genetics and evolutionary biology.[152] Because all the mitochondrial DNA is inherited as a single unit, or haplotype, the relationships between mitochondrial DNA from different individuals can be represented as a gene tree. Patterns in these gene trees can be used to infer the evolutionary history of populations. The classic example of this is in human evolutionary genetics, where the molecular clock can be used to provide a recent date for mitochondrial Eve.[153][154] This is often interpreted as strong support for a recent modern human expansion out of Africa.[155] Another human example is the sequencing of mitochondrial DNA from Neanderthal bones. The relatively large evolutionary distance between the mitochondrial DNA sequences of Neanderthals and living humans has been interpreted as evidence for the lack of interbreeding between Neanderthals and modern humans.[156]
However, mitochondrial DNA reflects only the history of the females in a population. This can be partially overcome by the use of paternal genetic sequences, such as the non-recombining region of the Y-chromosome.[155]
Recent measurements of the molecular clock for mitochondrial DNA[157] reported a value of 1 mutation every 7884 years dating back to the most recent common ancestor of humans and apes, which is consistent with estimates of mutation rates of autosomal DNA (10−8 per base per generation).[158]
Alternative genetic code
| Organism | Codon | Standard | Mitochondria |
|---|---|---|---|
| Mammals | AGA, AGG | Arginine | Stop codon |
| Invertebrates | AGA, AGG | Arginine | Serine |
| Fungi | CUA | Leucine | Threonine |
| All of the above | AUA | Isoleucine | Methionine |
| UGA | Stop codon | Tryptophan |
While slight variations on the standard genetic code had been predicted earlier,[159] none was discovered until 1979, when researchers studying human mitochondrial genes determined that they used an alternative code.[160] Nonetheless, the mitochondria of many other eukaryotes, including most plants, use the standard code.[161] Many slight variants have been discovered since,[161] including various alternative mitochondrial codes.[162] Further, the AUA, AUC, and AUU codons are all allowable start codons.
Some of these differences should be regarded as pseudo-changes in the genetic code due to the phenomenon of RNA editing, which is common in mitochondria. In higher plants, it was thought that CGG encoded for tryptophan and not arginine; however, the codon in the processed RNA was discovered to be the UGG codon, consistent with the standard genetic code for tryptophan.[163] Of note, the arthropod mitochondrial genetic code has undergone parallel evolution within a phylum, with some organisms uniquely translating AGG to lysine.[164]
Replication and inheritance
Mitochondria divide by mitochondrial fission, a form of binary fission that is also done by bacteria[165] although the process is tightly regulated by the host eukaryotic cell and involves communication between and contact with several other organelles. The regulation of this division differs between eukaryotes. In many single-celled eukaryotes, their growth and division are linked to the cell cycle. For example, a single mitochondrion may divide synchronously with the nucleus. This division and segregation process must be tightly controlled so that each daughter cell receives at least one mitochondrion. In other eukaryotes (in mammals for example), mitochondria may replicate their DNA and divide mainly in response to the energy needs of the cell, rather than in phase with the cell cycle. When the energy needs of a cell are high, mitochondria grow and divide. When energy use is low, mitochondria are destroyed or become inactive. In such examples mitochondria are apparently randomly distributed to the daughter cells during the division of the cytoplasm. Mitochondrial dynamics, the balance between mitochondrial fusion and fission, is an important factor in pathologies associated with several disease conditions.[166]
The hypothesis of mitochondrial binary fission has relied on the visualization by fluorescence microscopy and conventional transmission electron microscopy (TEM). The resolution of fluorescence microscopy (≈200 nm) is insufficient to distinguish structural details, such as double mitochondrial membrane in mitochondrial division or even to distinguish individual mitochondria when several are close together. Conventional TEM has also some technical limitations[which?] in verifying mitochondrial division. Cryo-electron tomography was recently used to visualize mitochondrial division in frozen hydrated intact cells. It revealed that mitochondria divide by budding.[167]
An individual's mitochondrial genes are inherited only from the mother, with rare exceptions.[168] In humans, when an egg cell is fertilized by a sperm, the mitochondria, and therefore the mitochondrial DNA, usually come from the egg only. The sperm's mitochondria enter the egg, but do not contribute genetic information to the embryo.[169] Instead, paternal mitochondria are marked with ubiquitin to select them for later destruction inside the embryo.[170] The egg cell contains relatively few mitochondria, but these mitochondria divide to populate the cells of the adult organism. This mode is seen in most organisms, including the majority of animals. However, mitochondria in some species can sometimes be inherited paternally. This is the norm among certain coniferous plants, although not in pine trees and yews.[171] For Mytilids, paternal inheritance only occurs within males of the species.[172][173][174] It has been suggested that it occurs at a very low level in humans.[175]
Uniparental inheritance leads to little opportunity for genetic recombination between different lineages of mitochondria, although a single mitochondrion can contain 2–10 copies of its DNA.[142] What recombination does take place maintains genetic integrity rather than maintaining diversity. However, there are studies showing evidence of recombination in mitochondrial DNA. It is clear that the enzymes necessary for recombination are present in mammalian cells.[176] Further, evidence suggests that animal mitochondria can undergo recombination.[177] The data are more controversial in humans, although indirect evidence of recombination exists.[178][179]
Entities undergoing uniparental inheritance and with little to no recombination may be expected to be subject to Muller's ratchet, the accumulation of deleterious mutations until functionality is lost. Animal populations of mitochondria avoid this buildup through a developmental process known as the mtDNA bottleneck. The bottleneck exploits stochastic processes in the cell to increase the cell-to-cell variability in mutant load as an organism develops: a single egg cell with some proportion of mutant mtDNA thus produces an embryo where different cells have different mutant loads. Cell-level selection may then act to remove those cells with more mutant mtDNA, leading to a stabilization or reduction in mutant load between generations. The mechanism underlying the bottleneck is debated,[180][181][182] with a recent mathematical and experimental metastudy providing evidence for a combination of random partitioning of mtDNAs at cell divisions and random turnover of mtDNA molecules within the cell.[183]
DNA repair
Mitochondria can repair oxidative DNA damage by mechanisms analogous to those occurring in the cell nucleus. The proteins employed in mtDNA repair are encoded by nuclear genes, and are translocated to the mitochondria. The DNA repair pathways in mammalian mitochondria include base excision repair, double-strand break repair, direct reversal and mismatch repair.[184][185] Alternatively, DNA damage may be bypassed, rather than repaired, by translesion synthesis.
Of the several DNA repair process in mitochondria, the base excision repair pathway has been most comprehensively studied.[185] Base excision repair is carried out by a sequence of enzyme-catalyzed steps that include recognition and excision of a damaged DNA base, removal of the resulting abasic site, end processing, gap filling and ligation. A common damage in mtDNA that is repaired by base excision repair is 8-oxoguanine produced by oxidation of guanine.[186]
Double-strand breaks can be repaired by homologous recombinational repair in both mammalian mtDNA[187] and plant mtDNA.[188] Double-strand breaks in mtDNA can also be repaired by microhomology-mediated end joining.[189] Although there is evidence for the repair processes of direct reversal and mismatch repair in mtDNA, these processes are not well characterized.[185]
Lack of mitochondrial DNA
Some organisms have lost mitochondrial DNA altogether. In these cases, genes encoded by the mitochondrial DNA have been lost or transferred to the nucleus.[141] Cryptosporidium have mitochondria that lack any DNA, presumably because all their genes have been lost or transferred.[190] In Cryptosporidium, the mitochondria have an altered ATP generation system that renders the parasite resistant to many classical mitochondrial inhibitors such as cyanide, azide, and atovaquone.[190] Mitochondria that lack their own DNA have been found in a marine parasitic dinoflagellate from the genus Amoebophyra. This microorganism, A. cerati, has functional mitochondria that lack a genome.[191] In related species, the mitochondrial genome still has three genes, but in A. cerati only a single mitochondrial gene — the cytochrome c oxidase I gene (cox1) — is found, and it has migrated to the genome of the nucleus.[192]
Dysfunction and disease
Mitochondrial diseases
Damage and subsequent dysfunction in mitochondria is an important factor in a range of human diseases due to their influence in cell metabolism. Mitochondrial disorders often present as neurological disorders, including autism.[18] They can also manifest as myopathy, diabetes, multiple endocrinopathy, and a variety of other systemic disorders.[193] Diseases caused by mutation in the mtDNA include Kearns–Sayre syndrome, MELAS syndrome and Leber's hereditary optic neuropathy.[194] In the vast majority of cases, these diseases are transmitted by a female to her children, as the zygote derives its mitochondria and hence its mtDNA from the ovum. Diseases such as Kearns-Sayre syndrome, Pearson syndrome, and progressive external ophthalmoplegia are thought to be due to large-scale mtDNA rearrangements, whereas other diseases such as MELAS syndrome, Leber's hereditary optic neuropathy, MERRF syndrome, and others are due to point mutations in mtDNA.[193]
It has also been reported that drug tolerant cancer cells have an increased number and size of mitochondria which suggested an increase in mitochondrial biogenesis.[195] A 2022 study in Nature Nanotechnology has reported that cancer cells can hijack the mitochondria from immune cells via physical tunneling nanotubes.[196]
In other diseases, defects in nuclear genes lead to dysfunction of mitochondrial proteins. This is the case in Friedreich's ataxia, hereditary spastic paraplegia, and Wilson's disease.[197] These diseases are inherited in a dominance relationship, as applies to most other genetic diseases. A variety of disorders can be caused by nuclear mutations of oxidative phosphorylation enzymes, such as coenzyme Q10 deficiency and Barth syndrome.[193] Environmental influences may interact with hereditary predispositions and cause mitochondrial disease. For example, there may be a link between pesticide exposure and the later onset of Parkinson's disease.[198][199] Other pathologies with etiology involving mitochondrial dysfunction include schizophrenia, bipolar disorder, dementia, Alzheimer's disease,[200][201] Parkinson's disease, epilepsy, stroke, cardiovascular disease, myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS), retinitis pigmentosa, and diabetes mellitus.[202][203]
Mitochondria-mediated oxidative stress plays a role in cardiomyopathy in type 2 diabetics. Increased fatty acid delivery to the heart increases fatty acid uptake by cardiomyocytes, resulting in increased fatty acid oxidation in these cells. This process increases the reducing equivalents available to the electron transport chain of the mitochondria, ultimately increasing reactive oxygen species (ROS) production. ROS increases uncoupling proteins (UCPs) and potentiate proton leakage through the adenine nucleotide translocator (ANT), the combination of which uncouples the mitochondria. Uncoupling then increases oxygen consumption by the mitochondria, compounding the increase in fatty acid oxidation. This creates a vicious cycle of uncoupling; furthermore, even though oxygen consumption increases, ATP synthesis does not increase proportionally because the mitochondria are uncoupled. Less ATP availability ultimately results in an energy deficit presenting as reduced cardiac efficiency and contractile dysfunction. To compound the problem, impaired sarcoplasmic reticulum calcium release and reduced mitochondrial reuptake limits peak cytosolic levels of the important signaling ion during muscle contraction. Decreased intra-mitochondrial calcium concentration increases dehydrogenase activation and ATP synthesis. So in addition to lower ATP synthesis due to fatty acid oxidation, ATP synthesis is impaired by poor calcium signaling as well, causing cardiac problems for diabetics.[204]
Mitochondria also modulate processes such as testicular somatic cell development, spermatogonial stem cell differentiation, luminal acidification, testosterone production in testes, and more. Thus, dysfunction of mitochondria in spermatozoa can be a cause for infertility.[205]
In efforts to combat mitochondrial disease, mitochondrial replacement therapy (MRT) has been developed. This form of in vitro fertilization uses donor mitochondria, which avoids the transmission of diseases caused by mutations of mitochondrial DNA.[206] However, this therapy is still being researched and can introduce genetic modification, as well as safety concerns. These diseases are rare but can be extremely debilitating and progressive diseases, thus posing complex ethical questions for public policy.[207]
Relationships to aging
There may be some leakage of the electrons transferred in the respiratory chain to form reactive oxygen species. This was thought to result in significant oxidative stress in the mitochondria with high mutation rates of mitochondrial DNA.[208] Hypothesized links between aging and oxidative stress are not new and were proposed in 1956,[209] which was later refined into the mitochondrial free radical theory of aging.[210] A vicious cycle was thought to occur, as oxidative stress leads to mitochondrial DNA mutations, which can lead to enzymatic abnormalities and further oxidative stress.
A number of changes can occur to mitochondria during the aging process.[211] Tissues from elderly humans show a decrease in enzymatic activity of the proteins of the respiratory chain.[212] However, mutated mtDNA can only be found in about 0.2% of very old cells.[213] Large deletions in the mitochondrial genome have been hypothesized to lead to high levels of oxidative stress and neuronal death in Parkinson's disease.[214] Mitochondrial dysfunction has also been shown to occur in amyotrophic lateral sclerosis.[215][216]
Since mitochondria cover a pivotal role in the ovarian function, by providing ATP necessary for the development from germinal vesicle to mature oocyte, a decreased mitochondria function can lead to inflammation, resulting in premature ovarian failure and accelerated ovarian aging. The resulting dysfunction is then reflected in quantitative (such as mtDNA copy number and mtDNA deletions), qualitative (such as mutations and strand breaks) and oxidative damage (such as dysfunctional mitochondria due to ROS), which are not only relevant in ovarian aging, but perturb oocyte-cumulus crosstalk in the ovary, are linked to genetic disorders (such as Fragile X) and can interfere with embryo selection.[217]
History
The first observations of intracellular structures that probably represented mitochondria were published in 1857, by the physiologist Albert von Kolliker.[218][219] Richard Altmann, in 1890, established them as cell organelles and called them "bioblasts".[219][220] In 1898, Carl Benda coined the term "mitochondria" from the Greek μίτος, mitos, "thread", and χονδρίον, chondrion, "granule".[221][219][222] Leonor Michaelis discovered that Janus green can be used as a supravital stain for mitochondria in 1900.[223] In 1904, Friedrich Meves made the first recorded observation of mitochondria in plants in cells of the white waterlily, Nymphaea alba,[219][224] and in 1908, along with Claudius Regaud, suggested that they contain proteins and lipids. Benjamin F. Kingsbury, in 1912, first related them with cell respiration, but almost exclusively based on morphological observations.[225][219] In 1913, Otto Heinrich Warburg linked respiration to particles which he had obtained from extracts of guinea-pig liver and which he called "grana".[226] Warburg and Heinrich Otto Wieland, who had also postulated a similar particle mechanism, disagreed on the chemical nature of the respiration. It was not until 1925, when David Keilin discovered cytochromes, that the respiratory chain was described.[219]
In 1939, experiments using minced muscle cells demonstrated that cellular respiration using one oxygen molecule can form four adenosine triphosphate (ATP) molecules, and in 1941, the concept of the phosphate bonds of ATP being a form of energy in cellular metabolism was developed by Fritz Albert Lipmann. In the following years, the mechanism behind cellular respiration was further elaborated, although its link to the mitochondria was not known.[219] The introduction of tissue fractionation by Albert Claude allowed mitochondria to be isolated from other cell fractions and biochemical analysis to be conducted on them alone. In 1946, he concluded that cytochrome oxidase and other enzymes responsible for the respiratory chain were isolated to the mitochondria. Eugene Kennedy and Albert Lehninger discovered in 1948 that mitochondria are the site of oxidative phosphorylation in eukaryotes. Over time, the fractionation method was further developed, improving the quality of the mitochondria isolated, and other elements of cell respiration were determined to occur in the mitochondria.[219]
The first high-resolution electron micrographs appeared in 1952, replacing the Janus Green stains as the preferred way to visualize mitochondria.[219] This led to a more detailed analysis of the structure of the mitochondria, including confirmation that they were surrounded by a membrane. It also showed a second membrane inside the mitochondria that folded up in ridges dividing up the inner chamber and that the size and shape of the mitochondria varied from cell to cell.
The popular term "powerhouse of the cell" was coined by Philip Siekevitz in 1957.[4][227]
In 1967, it was discovered that mitochondria contained ribosomes.[228] In 1968, methods were developed for mapping the mitochondrial genes, with the genetic and physical map of yeast mitochondrial DNA completed in 1976.[219] In November 2024, Researchers from the United States have discovered that mitochondria divide into two distinct forms when cells are starved, this could help explain and describe how cancers thrive in hostile conditions.https://www.nature.com/articles/d41586-024-03646-1
See also
References
- ^ "mitochondrion". Lexico UK English Dictionary. Oxford University Press. Archived from the original on January 2, 2020.
- ^ Campbell NA, Williamson B, Heyden RJ (2006). Biology: Exploring Life. Boston, Massachusetts: Pearson/Prentice Hall. ISBN 978-0132508827. Archived from the original on November 2, 2014. Retrieved January 6, 2009.
- ^ "Mighty Mitochondria and Neurodegenerative Diseases". Science in the News. February 1, 2012. Archived from the original on April 6, 2022. Retrieved April 24, 2022.
- ^ a b Siekevitz P (1957). "Powerhouse of the cell". Scientific American. 197 (1): 131–140. Bibcode:1957SciAm.197a.131S. doi:10.1038/scientificamerican0757-131.
- ^ a b c d Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, et al. (May 2016). "A Eukaryote without a Mitochondrial Organelle". Current Biology. 26 (10): 1274–1284. Bibcode:2016CBio...26.1274K. doi:10.1016/j.cub.2016.03.053. PMID 27185558.
- ^ Le Page M. "Animal that doesn't need oxygen to survive discovered New Scientist". New Scientist. Archived from the original on February 26, 2020. Retrieved February 25, 2020.
- ^ a b Yahalomi D, Atkinson SD, Neuhof M, Chang ES, Philippe H, Cartwright P, et al. (March 2020). "A cnidarian parasite of salmon (Myxozoa: Henneguya) lacks a mitochondrial genome". Proceedings of the National Academy of Sciences of the United States of America. 117 (10): 5358–5363. Bibcode:2020PNAS..117.5358Y. doi:10.1073/pnas.1909907117. PMC 7071853. PMID 32094163.
- ^ a b Henze K, Martin W (November 2003). "Evolutionary biology: essence of mitochondria". Nature. 426 (6963): 127–128. Bibcode:2003Natur.426..127H. doi:10.1038/426127a. PMID 14614484. S2CID 862398.
- ^ Leger MM, Kolisko M, Kamikawa R, Stairs CW, Kume K, Čepička I, et al. (April 2017). "Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes". Nature Ecology & Evolution. 1 (4): 0092. Bibcode:2017NatEE...1...92L. doi:10.1038/s41559-017-0092. PMC 5411260. PMID 28474007.
- ^ a b Novák LV, Treitli SC, Pyrih J, Hałakuc P, Pipaliya SV, Vacek V, et al. (December 2023). Dutcher SK (ed.). "Genomics of Preaxostyla Flagellates Illuminates the Path Towards the Loss of Mitochondria". PLOS Genetics. 19 (12): e1011050. doi:10.1371/journal.pgen.1011050. PMC 10703272. PMID 38060519.
- ^ Wiemerslage L, Lee D (March 2016). "Quantification of mitochondrial morphology in neurites of dopaminergic neurons using multiple parameters". Journal of Neuroscience Methods. 262: 56–65. doi:10.1016/j.jneumeth.2016.01.008. PMC 4775301. PMID 26777473.
- ^ a b c McBride HM, Neuspiel M, Wasiak S (July 2006). "Mitochondria: more than just a powerhouse". Current Biology. 16 (14): R551 – R560. Bibcode:2006CBio...16.R551M. doi:10.1016/j.cub.2006.06.054. PMID 16860735. S2CID 16252290.
- ^ Valero T (2014). "Mitochondrial biogenesis: pharmacological approaches". Current Pharmaceutical Design. 20 (35): 5507–5509. doi:10.2174/138161282035140911142118. hdl:10454/13341. PMID 24606795.
Mitochondrial biogenesis is therefore defined as the process via which cells increase their individual mitochondrial mass [3]. ... Mitochondrial biogenesis occurs by growth and division of pre-existing organelles and is temporally coordinated with cell cycle events [1].
- ^ Sanchis-Gomar F, García-Giménez JL, Gómez-Cabrera MC, Pallardó FV (2014). "Mitochondrial biogenesis in health and disease. Molecular and therapeutic approaches". Current Pharmaceutical Design. 20 (35): 5619–5633. doi:10.2174/1381612820666140306095106. PMID 24606801.
Mitochondrial biogenesis (MB) is the essential mechanism by which cells control the number of mitochondria
- ^ Gardner A, Boles RG (2005). "Is a 'Mitochondrial Psychiatry' in the Future? A Review". Curr. Psychiatry Rev. 1 (3): 255–271. doi:10.2174/157340005774575064.
- ^ Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL (June 2001). "Mitochondrial dysfunction in cardiac disease: ischemia--reperfusion, aging, and heart failure". Journal of Molecular and Cellular Cardiology. 33 (6): 1065–1089. doi:10.1006/jmcc.2001.1378. PMID 11444914.
- ^ Dorn GW, Vega RB, Kelly DP (October 2015). "Mitochondrial biogenesis and dynamics in the developing and diseased heart". Genes & Development. 29 (19): 1981–1991. doi:10.1101/gad.269894.115. PMC 4604339. PMID 26443844.
- ^ a b Griffiths KK, Levy RJ (2017). "Evidence of Mitochondrial Dysfunction in Autism: Biochemical Links, Genetic-Based Associations, and Non-Energy-Related Mechanisms". Oxidative Medicine and Cellular Longevity. 2017: 4314025. doi:10.1155/2017/4314025. PMC 5467355. PMID 28630658.
- ^ Ney PA (May 2011). "Normal and disordered reticulocyte maturation". Current Opinion in Hematology. 18 (3): 152–157. doi:10.1097/MOH.0b013e328345213e. PMC 3157046. PMID 21423015.
- ^ a b c d e f g h i j Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2005). Molecular Biology of the Cell. New York: Garland Publishing Inc. ISBN 978-0815341055.
- ^ a b c d e f g h i j k l Voet D, Voet JC, Pratt CW (2006). Fundamentals of Biochemistry (2nd ed.). John Wiley and Sons, Inc. pp. 547, 556. ISBN 978-0471214953.
- ^ Andersson SG, Karlberg O, Canbäck B, Kurland CG (January 2003). "On the origin of mitochondria: a genomics perspective". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 358 (1429): 165–77, discussion 177–9. doi:10.1098/rstb.2002.1193. PMC 1693097. PMID 12594925.
- ^ a b Gabaldón T (October 2021). "Origin and Early Evolution of the Eukaryotic Cell". Annual Review of Microbiology. 75 (1): 631–647. doi:10.1146/annurev-micro-090817-062213. PMID 34343017. S2CID 236916203.
- ^ "Mitochondrion – much more than an energy converter". British Society for Cell Biology. Archived from the original on April 4, 2019. Retrieved August 19, 2013.
- ^ Blachly-Dyson E, Forte M (September 2001). "VDAC channels". IUBMB Life. 52 (3–5): 113–118. doi:10.1080/15216540152845902. PMID 11798022. S2CID 38314888.
- ^ Hoogenboom BW, Suda K, Engel A, Fotiadis D (July 2007). "The supramolecular assemblies of voltage-dependent anion channels in the native membrane". Journal of Molecular Biology. 370 (2): 246–255. doi:10.1016/j.jmb.2007.04.073. PMID 17524423.
- ^ Zeth K (June 2010). "Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1797 (6–7): 1292–1299. doi:10.1016/j.bbabio.2010.04.019. PMID 20450883.
- ^ a b Herrmann JM, Neupert W (April 2000). "Protein transport into mitochondria" (PDF). Current Opinion in Microbiology. 3 (2): 210–214. doi:10.1016/S1369-5274(00)00077-1. PMID 10744987. Archived (PDF) from the original on August 16, 2022. Retrieved July 10, 2022.
- ^ a b Chipuk JE, Bouchier-Hayes L, Green DR (August 2006). "Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario". Cell Death and Differentiation. 13 (8): 1396–1402. doi:10.1038/sj.cdd.4401963. PMID 16710362.
- ^ a b c d e Hayashi T, Rizzuto R, Hajnoczky G, Su TP (February 2009). "MAM: more than just a housekeeper". Trends in Cell Biology. 19 (2): 81–88. doi:10.1016/j.tcb.2008.12.002. PMC 2750097. PMID 19144519.
- ^ Schenkel LC, Bakovic M (January 2014). "Formation and regulation of mitochondrial membranes". International Journal of Cell Biology. 2014: 709828. doi:10.1155/2014/709828. PMC 3918842. PMID 24578708.
- ^ McMillin JB, Dowhan W (December 2002). "Cardiolipin and apoptosis". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1585 (2–3): 97–107. doi:10.1016/S1388-1981(02)00329-3. PMID 12531542.
- ^ Bautista JS, Falabella M, Flannery PJ, Hanna MG, Heales SJ, Pope SA, et al. (December 2022). "Advances in methods to analyse cardiolipin and their clinical applications". Trends in Analytical Chemistry. 157: 116808. doi:10.1016/j.trac.2022.116808. PMC 7614147. PMID 36751553. S2CID 253211400.
- ^ Youle RJ, van der Bliek AM (August 2012). "Mitochondrial fission, fusion, and stress". Science. 337 (6098): 1062–1065. Bibcode:2012Sci...337.1062Y. doi:10.1126/science.1219855. PMC 4762028. PMID 22936770.
- ^ Cserép C, Pósfai B, Schwarcz AD, Dénes Á (2018). "Mitochondrial Ultrastructure Is Coupled to Synaptic Performance at Axonal Release Sites". eNeuro. 5 (1): ENEURO.0390–17.2018. doi:10.1523/ENEURO.0390-17.2018. PMC 5788698. PMID 29383328.
- ^ Mannella CA (2006). "Structure and dynamics of the mitochondrial inner membrane cristae". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1763 (5–6): 542–548. doi:10.1016/j.bbamcr.2006.04.006. PMID 16730811.
- ^ Bogenhagen DF (September 2012). "Mitochondrial DNA nucleoid structure". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1819 (9–10): 914–920. doi:10.1016/j.bbagrm.2011.11.005. PMID 22142616.
- ^ Rich PR (December 2003). "The molecular machinery of Keilin's respiratory chain". Biochemical Society Transactions. 31 (Pt 6): 1095–1105. doi:10.1042/BST0311095. PMID 14641005.
- ^ Stoimenova M, Igamberdiev AU, Gupta KJ, Hill RD (July 2007). "Nitrite-driven anaerobic ATP synthesis in barley and rice root mitochondria". Planta. 226 (2): 465–474. Bibcode:2007Plant.226..465S. doi:10.1007/s00425-007-0496-0. PMID 17333252. S2CID 8963850.
- ^ Neupert W (1997). "Protein import into mitochondria". Annual Review of Biochemistry. 66: 863–917. doi:10.1146/annurev.biochem.66.1.863. PMID 9242927.
- ^ a b c d e f Stryer L (1995). "Citric acid cycle.". In: Biochemistry (Fourth ed.). New York: W.H. Freeman and Company. pp. 509–527, 569–579, 614–616, 638–641, 732–735, 739–748, 770–773. ISBN 0716720094.
- ^ King A, Selak MA, Gottlieb E (August 2006). "Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer". Oncogene. 25 (34): 4675–4682. doi:10.1038/sj.onc.1209594. PMID 16892081. S2CID 26263513.
- ^ a b Voet D, Voet JG (2004). Biochemistry (3rd ed.). New York, NY: Wiley. p. 804. ISBN 978-0-471-19350-0.
- ^ a b Atkins P, de Paula J (2006). "Impact on biochemistry: Energy conversion in biological cells". Physical Chemistry (8th ed.). New York: Freeman. pp. 225–229. ISBN 978-0-7167-8759-4.
- ^ Huang H, Manton KG (May 2004). "The role of oxidative damage in mitochondria during aging: a review" (PDF). Frontiers in Bioscience. 9 (1–3): 1100–1117. doi:10.2741/1298. PMID 14977532. S2CID 2278219. Archived from the original (PDF) on March 3, 2019.
- ^ Mitchell P, Moyle J (January 1967). "Chemiosmotic hypothesis of oxidative phosphorylation". Nature. 213 (5072): 137–139. Bibcode:1967Natur.213..137M. doi:10.1038/213137a0. PMID 4291593. S2CID 4149605.
- ^ Mitchell P (June 1967). "Proton current flow in mitochondrial systems". Nature. 214 (5095): 1327–1328. Bibcode:1967Natur.214.1327M. doi:10.1038/2141327a0. PMID 6056845. S2CID 4160146.
- ^ Nobel Foundation. "Chemistry 1997". Archived from the original on July 8, 2007. Retrieved December 16, 2007.
- ^ a b Mozo J, Emre Y, Bouillaud F, Ricquier D, Criscuolo F (November 2005). "Thermoregulation: what role for UCPs in mammals and birds?". Bioscience Reports. 25 (3–4): 227–249. doi:10.1007/s10540-005-2887-4. PMID 16283555. S2CID 164450.
- ^ Kastaniotis AJ, Autio KJ, Kerätär JM, Monteuuis G, Mäkelä AM, Nair RR, et al. (January 2017). "Mitochondrial fatty acid synthesis, fatty acids and mitochondrial physiology". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1862 (1): 39–48. doi:10.1016/j.bbalip.2016.08.011. PMID 27553474.
- ^ Clay HB, Parl AK, Mitchell SL, Singh L, Bell LN, Murdock DG (March 2016). Peterson J (ed.). "Altering the Mitochondrial Fatty Acid Synthesis (mtFASII) Pathway Modulates Cellular Metabolic States and Bioactive Lipid Profiles as Revealed by Metabolomic Profiling". PLOS ONE. 11 (3): e0151171. Bibcode:2016PLoSO..1151171C. doi:10.1371/journal.pone.0151171. PMC 4786287. PMID 26963735.
- ^ a b c Nowinski SM, Van Vranken JG, Dove KK, Rutter J (October 2018). "Impact of Mitochondrial Fatty Acid Synthesis on Mitochondrial Biogenesis". Current Biology. 28 (20): R1212 – R1219. Bibcode:2018CBio...28R1212N. doi:10.1016/j.cub.2018.08.022. PMC 6258005. PMID 30352195.
- ^ Wehbe Z, Behringer S, Alatibi K, Watkins D, Rosenblatt D, Spiekerkoetter U, et al. (November 2019). "The emerging role of the mitochondrial fatty-acid synthase (mtFASII) in the regulation of energy metabolism". Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1864 (11): 1629–1643. doi:10.1016/j.bbalip.2019.07.012. PMID 31376476. S2CID 199404906.
- ^ Santulli G, Xie W, Reiken SR, Marks AR (September 2015). "Mitochondrial calcium overload is a key determinant in heart failure". Proceedings of the National Academy of Sciences of the United States of America. 112 (36): 11389–11394. Bibcode:2015PNAS..11211389S. doi:10.1073/pnas.1513047112. PMC 4568687. PMID 26217001.
- ^ a b Siegel GJ, Agranoff BW, Fisher SK, Albers RW, Uhler MD, eds. (1999). Basic Neurochemistry (6 ed.). Lippincott Williams & Wilkins. ISBN 978-0397518203.
- ^ a b Rossier MF (August 2006). "T channels and steroid biosynthesis: in search of a link with mitochondria". Cell Calcium. 40 (2): 155–164. doi:10.1016/j.ceca.2006.04.020. PMID 16759697.
- ^ Brighton CT, Hunt RM (May 1974). "Mitochondrial calcium and its role in calcification. Histochemical localization of calcium in electron micrographs of the epiphyseal growth plate with K-pyroantimonate". Clinical Orthopaedics and Related Research. 100 (5): 406–416. doi:10.1097/00003086-197405000-00057. PMID 4134194.
- ^ Brighton CT, Hunt RM (July 1978). "The role of mitochondria in growth plate calcification as demonstrated in a rachitic model". The Journal of Bone and Joint Surgery. American Volume. 60 (5): 630–639. doi:10.2106/00004623-197860050-00007. PMID 681381.
- ^ a b c Santulli G, Marks AR (2015). "Essential Roles of Intracellular Calcium Release Channels in Muscle, Brain, Metabolism, and Aging". Current Molecular Pharmacology. 8 (2): 206–222. doi:10.2174/1874467208666150507105105. PMID 25966694.
- ^ Pizzo P, Pozzan T (October 2007). "Mitochondria-endoplasmic reticulum choreography: structure and signaling dynamics". Trends in Cell Biology. 17 (10): 511–517. doi:10.1016/j.tcb.2007.07.011. PMID 17851078.
- ^ a b Miller RJ (March 1998). "Mitochondria - the Kraken wakes!". Trends in Neurosciences. 21 (3): 95–97. doi:10.1016/S0166-2236(97)01206-X. PMID 9530913. S2CID 5193821.
- ^ Santulli G, Pagano G, Sardu C, Xie W, Reiken S, D'Ascia SL, et al. (May 2015). "Calcium release channel RyR2 regulates insulin release and glucose homeostasis". The Journal of Clinical Investigation. 125 (5): 1968–1978. doi:10.1172/JCI79273. PMC 4463204. PMID 25844899.
- ^ Schwarzländer M, Logan DC, Johnston IG, Jones NS, Meyer AJ, Fricker MD, et al. (March 2012). "Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis". The Plant Cell. 24 (3): 1188–1201. Bibcode:2012PlanC..24.1188S. doi:10.1105/tpc.112.096438. PMC 3336130. PMID 22395486.
- ^ Ivannikov MV, Macleod GT (June 2013). "Mitochondrial free Ca²⁺ levels and their effects on energy metabolism in Drosophila motor nerve terminals". Biophysical Journal. 104 (11): 2353–2361. Bibcode:2013BpJ...104.2353I. doi:10.1016/j.bpj.2013.03.064. PMC 3672877. PMID 23746507.
- ^ Weinberg F, Chandel NS (October 2009). "Mitochondrial metabolism and cancer". Annals of the New York Academy of Sciences. 1177 (1): 66–73. Bibcode:2009NYASA1177...66W. doi:10.1111/j.1749-6632.2009.05039.x. PMID 19845608. S2CID 29827252.
- ^ a b Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E (March 2007). "Energy metabolism in tumor cells". The FEBS Journal. 274 (6): 1393–1418. doi:10.1111/j.1742-4658.2007.05686.x. PMID 17302740. S2CID 7748115.
- ^ Mistry JJ, Marlein CR, Moore JA, Hellmich C, Wojtowicz EE, Smith JG, et al. (December 2019). "ROS-mediated PI3K activation drives mitochondrial transfer from stromal cells to hematopoietic stem cells in response to infection". Proceedings of the National Academy of Sciences of the United States of America. 116 (49): 24610–24619. Bibcode:2019PNAS..11624610M. doi:10.1073/pnas.1913278116. PMC 6900710. PMID 31727843.
- ^ Pedersen PL (December 1994). "ATP synthase. The machine that makes ATP". Current Biology. 4 (12): 1138–1141. Bibcode:1994CBio....4.1138P. doi:10.1016/S0960-9822(00)00257-8. PMID 7704582. S2CID 10279742.
- ^ Pattappa G, Heywood HK, de Bruijn JD, Lee DA (October 2011). "The metabolism of human mesenchymal stem cells during proliferation and differentiation". Journal of Cellular Physiology. 226 (10): 2562–2570. doi:10.1002/jcp.22605. PMID 21792913. S2CID 22259833.
- ^ Agarwal B (June 2011). "A role for anions in ATP synthesis and its molecular mechanistic interpretation". Journal of Bioenergetics and Biomembranes. 43 (3): 299–310. doi:10.1007/s10863-011-9358-3. PMID 21647635. S2CID 29715383.
- ^ a b c Sweet S, Singh G (July 1999). "Changes in mitochondrial mass, membrane potential, and cellular adenosine triphosphate content during the cell cycle of human leukemic (HL-60) cells". Journal of Cellular Physiology. 180 (1): 91–96. doi:10.1002/(SICI)1097-4652(199907)180:1<91::AID-JCP10>3.0.CO;2-6. PMID 10362021. S2CID 25906279.
- ^ Riera Romo M (August 2021). "Cell death as part of innate immunity: Cause or consequence?". Immunology. 163 (4): 399–415. doi:10.1111/imm.13325. PMC 8274179. PMID 33682112.
- ^ Green DR (September 1998). "Apoptotic pathways: the roads to ruin". Cell. 94 (6): 695–698. doi:10.1016/S0092-8674(00)81728-6. PMID 9753316. S2CID 16654082.
- ^ a b c d e f Bahat A, MacVicar T, Langer T (July 27, 2021). "Metabolism and Innate Immunity Meet at the Mitochondria". Frontiers in Cell and Developmental Biology. 9: 720490. doi:10.3389/fcell.2021.720490. PMC 8353256. PMID 34386501.
- ^ a b c d e f Murphy MP, O'Neill LA (February 2024). "A break in mitochondrial endosymbiosis as a basis for inflammatory diseases". Nature. 626 (7998): 271–279. Bibcode:2024Natur.626..271M. doi:10.1038/s41586-023-06866-z. PMID 38326590.
- ^ Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, et al. (April 2011). "Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation". Trends in Immunology. 32 (4): 157–164. doi:10.1016/j.it.2011.01.005. PMID 21334975.; cited in [75]
- ^ Riley JS, Tait SW (April 2020). "Mitochondrial DNA in inflammation and immunity". EMBO Reports. 21 (4): e49799. doi:10.15252/embr.201949799. PMC 7132203. PMID 32202065.; cited in [75]
- ^ Seth RB, Sun L, Ea CK, Chen ZJ (September 2005). "Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3". Cell. 122 (5): 669–682. doi:10.1016/j.cell.2005.08.012. PMID 16125763.; cited in [75]
- ^ Dorward DA, Lucas CD, Doherty MK, Chapman GB, Scholefield EJ, Conway Morris A, et al. (October 2017). "Novel role for endogenous mitochondrial formylated peptide-driven formyl peptide receptor 1 signalling in acute respiratory distress syndrome". Thorax. 72 (10): 928–936. doi:10.1136/thoraxjnl-2017-210030. PMC 5738532. PMID 28469031.; cited in [75]
- ^ Cai N, Gomez-Duran A, Yonova-Doing E, Kundu K, Burgess AI, Golder ZJ, et al. (September 2021). "Mitochondrial DNA variants modulate N-formylmethionine, proteostasis and risk of late-onset human diseases". Nature Medicine. 27 (9): 1564–1575. doi:10.1038/s41591-021-01441-3. PMID 34426706.; cited in [75]
- ^ Zhang W, Wang G, Xu ZG, Tu H, Hu F, Dai J, et al. (June 2019). "Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS". Cell. 178 (1): 176–189.e15. doi:10.1016/j.cell.2019.05.003. PMC 6625351. PMID 31155231.; cited in [74]
- ^ Pourcelot M, Arnoult D (September 2014). "Mitochondrial dynamics and the innate antiviral immune response". The FEBS Journal. 281 (17): 3791–3802. doi:10.1111/febs.12940. PMID 25051991.; cited in [74]
- ^ Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF (February 2013). "Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers". Journal of Hematology & Oncology. 6 (19): 19. doi:10.1186/1756-8722-6-19. PMC 3599349. PMID 23442817.
- ^ Hajnóczky G, Csordás G, Das S, Garcia-Perez C, Saotome M, Sinha Roy S, et al. (2006). "Mitochondrial calcium signalling and cell death: approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis". Cell Calcium. 40 (5–6): 553–560. doi:10.1016/j.ceca.2006.08.016. PMC 2692319. PMID 17074387.
- ^ Oh-hama T (August 1997). "Evolutionary consideration on 5-aminolevulinate synthase in nature". Origins of Life and Evolution of the Biosphere. 27 (4): 405–412. Bibcode:1997OLEB...27..405O. doi:10.1023/A:1006583601341. PMID 9249985. S2CID 13602877.
- ^ Klinge CM (December 2008). "Estrogenic control of mitochondrial function and biogenesis". Journal of Cellular Biochemistry. 105 (6): 1342–1351. doi:10.1002/jcb.21936. PMC 2593138. PMID 18846505.
- ^ Alvarez-Delgado C, Mendoza-Rodríguez CA, Picazo O, Cerbón M (August 2010). "Different expression of alpha and beta mitochondrial estrogen receptors in the aging rat brain: interaction with respiratory complex V". Experimental Gerontology. 45 (7–8): 580–585. doi:10.1016/j.exger.2010.01.015. PMID 20096765. S2CID 30841790.
- ^ Pavón N, Martínez-Abundis E, Hernández L, Gallardo-Pérez JC, Alvarez-Delgado C, Cerbón M, et al. (October 2012). "Sexual hormones: effects on cardiac and mitochondrial activity after ischemia-reperfusion in adult rats. Gender difference". The Journal of Steroid Biochemistry and Molecular Biology. 132 (1–2): 135–146. doi:10.1016/j.jsbmb.2012.05.003. PMID 22609314. S2CID 24794040.
- ^ Breda CN, Davanzo GG, Basso PJ, Saraiva Câmara NO, Moraes-Vieira PM (September 2019). "Mitochondria as central hub of the immune system". Redox Biology. 26: 101255. doi:10.1016/j.redox.2019.101255. PMC 6598836. PMID 31247505.
- ^ a b Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, et al. (January 2020). "Microglia monitor and protect neuronal function through specialized somatic purinergic junctions". Science. 367 (6477): 528–537. Bibcode:2020Sci...367..528C. doi:10.1126/science.aax6752. PMID 31831638. S2CID 209343260. Archived from the original on May 2, 2023. Retrieved February 10, 2022.
- ^ Cserép C, Schwarcz AD, Pósfai B, László ZI, Kellermayer A, Környei Z, et al. (September 2022). "Microglial control of neuronal development via somatic purinergic junctions". Cell Reports. 40 (12): 111369. doi:10.1016/j.celrep.2022.111369. PMC 9513806. PMID 36130488. S2CID 252416407.
- ^ Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, et al. (March 2003). "Characterization of the human heart mitochondrial proteome". Nature Biotechnology. 21 (3): 281–286. doi:10.1038/nbt793. PMID 12592411. S2CID 27329521.
- ^ Zhang J, Li X, Mueller M, Wang Y, Zong C, Deng N, et al. (April 2008). "Systematic characterization of the murine mitochondrial proteome using functionally validated cardiac mitochondria". Proteomics. 8 (8): 1564–1575. doi:10.1002/pmic.200700851. PMC 2799225. PMID 18348319.
- ^ Zhang J, Liem DA, Mueller M, Wang Y, Zong C, Deng N, et al. (June 2008). "Altered proteome biology of cardiac mitochondria under stress conditions". Journal of Proteome Research. 7 (6): 2204–2214. doi:10.1021/pr070371f. PMC 3805274. PMID 18484766.
- ^ Logan DC (June 2010). "Mitochondrial fusion, division and positioning in plants". Biochemical Society Transactions. 38 (3): 789–795. doi:10.1042/bst0380789. PMID 20491666.
- ^ das Neves RP, Jones NS, Andreu L, Gupta R, Enver T, Iborra FJ (December 2010). Weissman JS (ed.). "Connecting variability in global transcription rate to mitochondrial variability". PLOS Biology. 8 (12): e1000560. doi:10.1371/journal.pbio.1000560. PMC 3001896. PMID 21179497.
- ^ Johnston IG, Gaal B, Neves RP, Enver T, Iborra FJ, Jones NS (2012). Haugh JM (ed.). "Mitochondrial variability as a source of extrinsic cellular noise". PLOS Computational Biology. 8 (3): e1002416. arXiv:1107.4499. Bibcode:2012PLSCB...8E2416J. doi:10.1371/journal.pcbi.1002416. PMC 3297557. PMID 22412363.
- ^ Rappaport L, Oliviero P, Samuel JL (July 1998). "Cytoskeleton and mitochondrial morphology and function". Molecular and Cellular Biochemistry. 184 (1–2): 101–105. doi:10.1023/A:1006843113166. PMID 9746315. S2CID 28165195.
- ^ Hoitzing H, Johnston IG, Jones NS (June 2015). "What is the function of mitochondrial networks? A theoretical assessment of hypotheses and proposal for future research". BioEssays. 37 (6): 687–700. doi:10.1002/bies.201400188. PMC 4672710. PMID 25847815.
- ^ Soltys BJ, Gupta RS (1992). "Interrelationships of endoplasmic reticulum, mitochondria, intermediate filaments, and microtubules--a quadruple fluorescence labeling study". Biochemistry and Cell Biology. 70 (10–11): 1174–1186. doi:10.1139/o92-163. PMID 1363623.
- ^ Tang HL, Lung HL, Wu KC, Le AH, Tang HM, Fung MC (February 2008). "Vimentin supports mitochondrial morphology and organization". The Biochemical Journal. 410 (1): 141–146. doi:10.1042/BJ20071072. PMID 17983357.
- ^ a b c d e f g h i j k l m n Rizzuto R, Marchi S, Bonora M, Aguiari P, Bononi A, De Stefani D, et al. (November 2009). "Ca(2+) transfer from the ER to mitochondria: when, how and why". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1787 (11): 1342–1351. doi:10.1016/j.bbabio.2009.03.015. PMC 2730423. PMID 19341702.
- ^ a b de Brito OM, Scorrano L (August 2010). "An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship". The EMBO Journal. 29 (16): 2715–2723. doi:10.1038/emboj.2010.177. PMC 2924651. PMID 20717141.
- ^ a b Vance JE, Shiao YJ (1996). "Intracellular trafficking of phospholipids: import of phosphatidylserine into mitochondria". Anticancer Research. 16 (3B): 1333–1339. PMID 8694499.
- ^ a b c Lebiedzinska M, Szabadkai G, Jones AW, Duszynski J, Wieckowski MR (October 2009). "Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles". The International Journal of Biochemistry & Cell Biology. 41 (10): 1805–1816. doi:10.1016/j.biocel.2009.02.017. PMID 19703651.
- ^ Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. (January 2008). "Fission and selective fusion govern mitochondrial segregation and elimination by autophagy". The EMBO Journal. 27 (2): 433–446. doi:10.1038/sj.emboj.7601963. PMC 2234339. PMID 18200046.
- ^ a b c d e f Osman C, Voelker DR, Langer T (January 2011). "Making heads or tails of phospholipids in mitochondria". The Journal of Cell Biology. 192 (1): 7–16. doi:10.1083/jcb.201006159. PMC 3019561. PMID 21220505.
- ^ Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, et al. (July 2009). "An ER-mitochondria tethering complex revealed by a synthetic biology screen". Science. 325 (5939): 477–481. Bibcode:2009Sci...325..477K. doi:10.1126/science.1175088. PMC 2933203. PMID 19556461.
- ^ Rusiñol AE, Cui Z, Chen MH, Vance JE (November 1994). "A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins". The Journal of Biological Chemistry. 269 (44): 27494–27502. doi:10.1016/S0021-9258(18)47012-3. PMID 7961664.
- ^ a b Kopach O, Kruglikov I, Pivneva T, Voitenko N, Fedirko N (May 2008). "Functional coupling between ryanodine receptors, mitochondria and Ca(2+) ATPases in rat submandibular acinar cells". Cell Calcium. 43 (5): 469–481. doi:10.1016/j.ceca.2007.08.001. PMID 17889347.
- ^ Csordás G, Hajnóczky G (April 2001). "Sorting of calcium signals at the junctions of endoplasmic reticulum and mitochondria". Cell Calcium. 29 (4): 249–262. doi:10.1054/ceca.2000.0191. PMID 11243933.
- ^ a b c d Decuypere JP, Monaco G, Bultynck G, Missiaen L, De Smedt H, Parys JB (May 2011). "The IP(3) receptor-mitochondria connection in apoptosis and autophagy". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1813 (5): 1003–1013. doi:10.1016/j.bbamcr.2010.11.023. PMID 21146562.
- ^ Diercks BP, Fliegert R, Guse AH (June 2017). "Mag-Fluo4 in T cells: Imaging of intra-organelle free Ca2+ concentrations". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1864 (6): 977–986. doi:10.1016/j.bbamcr.2016.11.026. PMID 27913206.
- ^ Hajnóczky G, Csordás G, Yi M (2011). "Old players in a new role: mitochondria-associated membranes, VDAC, and ryanodine receptors as contributors to calcium signal propagation from endoplasmic reticulum to the mitochondria". Cell Calcium. 32 (5–6): 363–377. doi:10.1016/S0143416002001872. PMID 12543096.
- ^ Marriott KS, Prasad M, Thapliyal V, Bose HS (December 2012). "σ-1 receptor at the mitochondrial-associated endoplasmic reticulum membrane is responsible for mitochondrial metabolic regulation". The Journal of Pharmacology and Experimental Therapeutics. 343 (3): 578–586. doi:10.1124/jpet.112.198168. PMC 3500540. PMID 22923735.
- ^ Hayashi T, Su TP (November 2007). "Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival". Cell. 131 (3): 596–610. doi:10.1016/j.cell.2007.08.036. PMID 17981125. S2CID 18885068.
- ^ Csordás G, Weaver D, Hajnóczky G (July 2018). "Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions". Trends in Cell Biology. 28 (7): 523–540. doi:10.1016/j.tcb.2018.02.009. PMC 6005738. PMID 29588129.
- ^ McCutcheon JP (October 2021). "The Genomics and Cell Biology of Host-Beneficial Intracellular Infections". Annual Review of Cell and Developmental Biology. 37 (1): 115–142. doi:10.1146/annurev-cellbio-120219-024122. PMID 34242059. S2CID 235786110.
- ^ Callier V (June 8, 2022). "Mitochondria and the origin of eukaryotes". Knowable Magazine. doi:10.1146/knowable-060822-2. S2CID 249526889. Archived from the original on August 15, 2022. Retrieved August 18, 2022.
- ^ a b Margulis L, Sagan D (1986). Origins of Sex: Three billion years of genetic recombination. New Haven, CT: Yale University Press. pp. 69–71, 87. ISBN 978-0300033403.
- ^ Martin WF, Müller M (2007). Origin of mitochondria and hydrogenosomes. Heidelberg, DE: Springer Verlag.
- ^ Emelyanov VV (April 2003). "Mitochondrial connection to the origin of the eukaryotic cell". European Journal of Biochemistry. 270 (8): 1599–1618. doi:10.1046/j.1432-1033.2003.03499.x. PMID 12694174.
- ^ Müller M, Martin W (May 1999). "The genome of Rickettsia prowazekii and some thoughts on the origin of mitochondria and hydrogenosomes". BioEssays. 21 (5): 377–381. doi:10.1002/(sici)1521-1878(199905)21:5<377::aid-bies4>3.0.co;2-w. PMID 10376009.
- ^ Gray MW, Burger G, Lang BF (March 1999). "Mitochondrial evolution". Science. 283 (5407): 1476–1481. Bibcode:1999Sci...283.1476G. doi:10.1126/science.283.5407.1476. PMC 3428767. PMID 10066161.
- ^ Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, et al. (June 14, 2011). "Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade". Scientific Reports. 1 (1): 13. Bibcode:2011NatSR...1...13T. doi:10.1038/srep00013. PMC 3216501. PMID 22355532.
- ^ Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJ (May 2018). "Deep mitochondrial origin outside the sampled alphaproteobacteria". Nature. 557 (7703): 101–105. Bibcode:2018Natur.557..101M. doi:10.1038/s41586-018-0059-5. hdl:1874/373336. PMID 29695865. S2CID 13740626.
- ^ Fan L, Wu D, Goremykin V, Xiao J, Xu Y, Garg S, et al. (September 2020). "Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria". Nature Ecology & Evolution. 4 (9). Springer Science and Business Media LLC: 1213–1219. Bibcode:2020NatEE...4.1213F. bioRxiv 10.1101/715870. doi:10.1038/s41559-020-1239-x. PMID 32661403. S2CID 220507452.
- ^ Wang S, Luo H (June 2021). "Dating Alphaproteobacteria evolution with eukaryotic fossils". Nature Communications. 12 (1): 3324. Bibcode:2021NatCo..12.3324W. doi:10.1038/s41467-021-23645-4. PMC 8175736. PMID 34083540.
- ^ Esposti MD, Geiger O, Sanchez-Flores A (May 16, 2022). "On the bacterial ancestry of mitochondria: New insights with triangulated approaches". bioRxiv: 2022.05.15.491939. doi:10.1101/2022.05.15.491939. S2CID 248836055.
- ^ Muñoz-Gómez SA, Susko E, Williamson K, Eme L, Slamovits CH, Moreira D, et al. (March 2022). "Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria" (PDF). Nature Ecology & Evolution. 6 (3): 253–262. Bibcode:2022NatEE...6..253M. doi:10.1038/s41559-021-01638-2. PMID 35027725. S2CID 245958471. Archived (PDF) from the original on November 18, 2023. Retrieved December 30, 2023.
- ^ Schön ME, Martijn J, Vosseberg J, Köstlbacher S, Ettema TJ (August 2022). "The evolutionary origin of host association in the Rickettsiales". Nature Microbiology. 7 (8): 1189–1199. doi:10.1038/s41564-022-01169-x. PMC 9352585. PMID 35798888.
- ^ O'Brien TW (September 2003). "Properties of human mitochondrial ribosomes". IUBMB Life. 55 (9): 505–513. doi:10.1080/15216540310001626610. PMID 14658756. S2CID 43415449.
- ^ Sagan L (March 1967). "On the origin of mitosing cells". Journal of Theoretical Biology. 14 (3): 255–274. Bibcode:1967JThBi..14..225S. doi:10.1016/0022-5193(67)90079-3. PMID 11541392.
- ^ Emelyanov VV (February 2001). "Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria". Bioscience Reports. 21 (1): 1–17. doi:10.1023/A:1010409415723. PMID 11508688. S2CID 10144020.
- ^ Feng DF, Cho G, Doolittle RF (November 1997). "Determining divergence times with a protein clock: update and reevaluation". Proceedings of the National Academy of Sciences of the United States of America. 94 (24): 13028–13033. Bibcode:1997PNAS...9413028F. doi:10.1073/pnas.94.24.13028. PMC 24257. PMID 9371794.
- ^ Cavalier-Smith T (1991). "Archamoebae: the ancestral eukaryotes?". Bio Systems. 25 (1–2): 25–38. Bibcode:1991BiSys..25...25C. doi:10.1016/0303-2647(91)90010-I. PMID 1854912.
- ^ Danovaro R, Dell'Anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM (April 2010). "The first metazoa living in permanently anoxic conditions". BMC Biology. 8: 30. doi:10.1186/1741-7007-8-30. PMC 2907586. PMID 20370908.
- ^ Coghaln A (April 7, 2010). "The mud creature that lives without oxygen". Zoologger. New Scientist. Archived from the original on February 26, 2020. Retrieved February 27, 2020.
- ^ Shiflett AM, Johnson PJ (2010). "Mitochondrion-related organelles in eukaryotic protists". Annual Review of Microbiology. 64: 409–429. doi:10.1146/annurev.micro.62.081307.162826. PMC 3208401. PMID 20528687.
- ^ Karnkowska A, Treitli SC, Brzoň O, Novák L, Vacek V, Soukal P, et al. (October 2019). Battistuzzi FU (ed.). "The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion". Molecular Biology and Evolution. 36 (10): 2292–2312. doi:10.1093/molbev/msz147. PMC 6759080. PMID 31387118. Archived from the original on April 12, 2024. Retrieved April 14, 2024.
- ^ a b c Chan DC (June 2006). "Mitochondria: dynamic organelles in disease, aging, and development". Cell. 125 (7): 1241–1252. doi:10.1016/j.cell.2006.06.010. PMID 16814712. S2CID 8551160.
- ^ a b Wiesner RJ, Rüegg JC, Morano I (March 1992). "Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues". Biochemical and Biophysical Research Communications. 183 (2): 553–559. doi:10.1016/0006-291X(92)90517-O. PMID 1550563.
- ^ Falkenberg M (July 2018). "Mitochondrial DNA replication in mammalian cells: overview of the pathway". Essays in Biochemistry. 62 (3): 287–296. doi:10.1042/ebc20170100. PMC 6056714. PMID 29880722.
- ^ a b Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. (April 1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–465. Bibcode:1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534. S2CID 4355527.
- ^ Fukuhara H, Sor F, Drissi R, Dinouël N, Miyakawa I, Rousset S, et al. (April 1993). "Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features". Molecular and Cellular Biology. 13 (4): 2309–2314. doi:10.1128/mcb.13.4.2309. PMC 359551. PMID 8455612.
- ^ Bernardi G (December 1978). "Intervening sequences in the mitochondrial genome". Nature. 276 (5688): 558–559. Bibcode:1978Natur.276..558B. doi:10.1038/276558a0. PMID 214710. S2CID 4314378.
- ^ Hebbar SK, Belcher SM, Perlman PS (April 1992). "A maturase-encoding group IIA intron of yeast mitochondria self-splices in vitro". Nucleic Acids Research. 20 (7): 1747–1754. doi:10.1093/nar/20.7.1747. PMC 312266. PMID 1579468.
- ^ Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, et al. (February 1998). "Genome structure and gene content in protist mitochondrial DNAs". Nucleic Acids Research. 26 (4): 865–878. doi:10.1093/nar/26.4.865. PMC 147373. PMID 9461442.
- ^ Gray MW, Lang BF, Burger G (2004). "Mitochondria of protists". Annual Review of Genetics. 38: 477–524. doi:10.1146/annurev.genet.37.110801.142526. PMID 15568984.
- ^ Telonis AG, Loher P, Kirino Y, Rigoutsos I (2014). "Nuclear and mitochondrial tRNA-lookalikes in the human genome". Frontiers in Genetics. 5: 344. doi:10.3389/fgene.2014.00344. PMC 4189335. PMID 25339973.
- ^ Shao R, Kirkness EF, Barker SC (May 2009). "The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus". Genome Research. 19 (5): 904–912. doi:10.1101/gr.083188.108. PMC 2675979. PMID 19336451.
- ^ Castro JA, Picornell A, Ramon M (December 1998). "Mitochondrial DNA: a tool for populational genetics studies". International Microbiology. 1 (4): 327–332. PMID 10943382.
- ^ Cann RL, Stoneking M, Wilson AC (January 1987). "Mitochondrial DNA and human evolution". Nature. 325 (6099): 31–36. Bibcode:1987Natur.325...31C. doi:10.1038/325031a0. PMID 3025745. S2CID 4285418.
- ^ Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ (June 2006). "Harvesting the fruit of the human mtDNA tree". Trends in Genetics. 22 (6): 339–345. doi:10.1016/j.tig.2006.04.001. PMID 16678300.
- ^ a b Garrigan D, Hammer MF (September 2006). "Reconstructing human origins in the genomic era". Nature Reviews. Genetics. 7 (9): 669–680. doi:10.1038/nrg1941. PMID 16921345. S2CID 176541.
- ^ Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S (July 1997). "Neandertal DNA sequences and the origin of modern humans". Cell. 90 (1): 19–30. doi:10.1016/S0092-8674(00)80310-4. hdl:11858/00-001M-0000-0025-0960-8. PMID 9230299. S2CID 13581775.
- ^ Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, et al. (June 2009). "Correcting for purifying selection: an improved human mitochondrial molecular clock". American Journal of Human Genetics. 84 (6): 740–759. doi:10.1016/j.ajhg.2009.05.001. PMC 2694979. PMID 19500773.
- ^ Nachman MW, Crowell SL (September 2000). "Estimate of the mutation rate per nucleotide in humans". Genetics. 156 (1): 297–304. doi:10.1093/genetics/156.1.297. PMC 1461236. PMID 10978293.
- ^ Crick FH, Orgel LE (1973). "Directed panspermia" (PDF). Icarus. 19 (3): 341–346. Bibcode:1973Icar...19..341C. doi:10.1016/0019-1035(73)90110-3. Archived (PDF) from the original on October 29, 2011. Retrieved October 21, 2014.
p. 344: It is a little surprising that organisms with somewhat different codes do not coexist.
Further discussion Archived September 3, 2011, at the Wayback Machine. - ^ Barrell BG, Bankier AT, Drouin J (November 1979). "A different genetic code in human mitochondria". Nature. 282 (5735): 189–194. Bibcode:1979Natur.282..189B. doi:10.1038/282189a0. PMID 226894. S2CID 4335828.
- ^ a b Elzanowski A, Ostell J (January 7, 2019). "The Genetic Codes". www.ncbi.nlm.nih.gov. Archived from the original on May 13, 2011. Retrieved February 10, 2023.
- ^ Jukes TH, Osawa S (December 1990). "The genetic code in mitochondria and chloroplasts". Experientia. 46 (11–12): 1117–1126. doi:10.1007/BF01936921. PMID 2253709. S2CID 19264964.
- ^ Hiesel R, Wissinger B, Schuster W, Brennicke A (December 1989). "RNA editing in plant mitochondria". Science. 246 (4937): 1632–1634. Bibcode:1989Sci...246.1632H. doi:10.1126/science.2480644. PMID 2480644.
- ^ Abascal F, Posada D, Knight RD, Zardoya R (May 2006). "Parallel evolution of the genetic code in arthropod mitochondrial genomes". PLOS Biology. 4 (5): e127. doi:10.1371/journal.pbio.0040127. PMC 1440934. PMID 16620150.
- ^ Pfeiffer RF (2012). Parkinson's Disease. CRC Press. p. 583. ISBN 978-1439807149.
- ^ Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C (August 2010). "New insights into the role of mitochondria in aging: mitochondrial dynamics and more". Journal of Cell Science. 123 (Pt 15): 2533–2542. doi:10.1242/jcs.070490. PMC 2912461. PMID 20940129.
- ^ Hu GB (August 2014). "Whole cell cryo-electron tomography suggests mitochondria divide by budding". Microscopy and Microanalysis. 20 (4): 1180–1187. Bibcode:2014MiMic..20.1180H. doi:10.1017/S1431927614001317. PMID 24870811. S2CID 7440669.
- ^ McWilliams TG, Suomalainen A (January 2019). "Mitochondrial DNA can be inherited from fathers, not just mothers". Nature. 565 (7739): 296–297. Bibcode:2019Natur.565..296M. doi:10.1038/d41586-019-00093-1. PMID 30643304.
- ^ Kimball, J.W. (2006) "Sexual Reproduction in Humans: Copulation and Fertilization" Archived October 2, 2015, at the Wayback Machine, Kimball's Biology Pages (based on Biology, 6th ed., 1996)
- ^ Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G (November 1999). "Ubiquitin tag for sperm mitochondria". Nature. 402 (6760): 371–372. Bibcode:1999Natur.402..371S. doi:10.1038/46466. PMID 10586873. S2CID 205054671. Discussed in Science News Archived December 19, 2007, at the Wayback Machine.
- ^ Mogensen HL (1996). "The Hows and Whys of Cytoplasmic Inheritance in Seed Plants". American Journal of Botany. 83 (3): 383–404. doi:10.2307/2446172. JSTOR 2446172.
- ^ Zouros E (December 2000). "The exceptional mitochondrial DNA system of the mussel family Mytilidae". Genes & Genetic Systems. 75 (6): 313–318. doi:10.1266/ggs.75.313. PMID 11280005.
- ^ Sutherland B, Stewart D, Kenchington ER, Zouros E (January 1998). "The fate of paternal mitochondrial DNA in developing female mussels, Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA". Genetics. 148 (1): 341–347. doi:10.1093/genetics/148.1.341. PMC 1459795. PMID 9475744.
- ^ Male and Female Mitochondrial DNA Lineages in the Blue Mussel (Mytilus edulis) Species Group Archived May 18, 2013, at the Wayback Machine by Donald T. Stewart, Carlos Saavedra, Rebecca R. Stanwood, Amy 0. Ball, and Eleftherios Zouros
- ^ Johns DR (October 2003). "Paternal transmission of mitochondrial DNA is (fortunately) rare". Annals of Neurology. 54 (4): 422–424. doi:10.1002/ana.10771. PMID 14520651. S2CID 90422.
- ^ Thyagarajan B, Padua RA, Campbell C (November 1996). "Mammalian mitochondria possess homologous DNA recombination activity". The Journal of Biological Chemistry. 271 (44): 27536–27543. doi:10.1074/jbc.271.44.27536. PMID 8910339.
- ^ Lunt DH, Hyman BC (May 1997). "Animal mitochondrial DNA recombination". Nature. 387 (6630): 247. Bibcode:1997Natur.387..247L. doi:10.1038/387247a0. PMID 9153388. S2CID 4318161.
- ^ Eyre-Walker A, Smith NH, Smith JM (March 1999). "How clonal are human mitochondria?". Proceedings. Biological Sciences. 266 (1418): 477–483. doi:10.1098/rspb.1999.0662. PMC 1689787. PMID 10189711.
- ^ Awadalla P, Eyre-Walker A, Smith JM (December 1999). "Linkage disequilibrium and recombination in hominid mitochondrial DNA". Science. 286 (5449): 2524–2525. doi:10.1126/science.286.5449.2524. PMID 10617471. S2CID 27685285.
- ^ Cree LM, Samuels DC, de Sousa Lopes SC, Rajasimha HK, Wonnapinij P, Mann JR, et al. (February 2008). "A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes". Nature Genetics. 40 (2): 249–254. doi:10.1038/ng.2007.63. PMID 18223651. S2CID 205344980.
- ^ Cao L, Shitara H, Horii T, Nagao Y, Imai H, Abe K, et al. (March 2007). "The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells". Nature Genetics. 39 (3): 386–390. doi:10.1038/ng1970. PMID 17293866. S2CID 10686347.
- ^ Wai T, Teoli D, Shoubridge EA (December 2008). "The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes". Nature Genetics. 40 (12): 1484–1488. doi:10.1038/ng.258. PMID 19029901. S2CID 225349.
- ^ Johnston IG, Burgstaller JP, Havlicek V, Kolbe T, Rülicke T, Brem G, et al. (June 2015). "Stochastic modelling, Bayesian inference, and new in vivo measurements elucidate the debated mtDNA bottleneck mechanism". eLife. 4: e07464. arXiv:1512.02988. doi:10.7554/eLife.07464. PMC 4486817. PMID 26035426.
- ^ Gredilla R, Garm C, Stevnsner T (2012). "Nuclear and mitochondrial DNA repair in selected eukaryotic aging model systems". Oxidative Medicine and Cellular Longevity. 2012: 282438. doi:10.1155/2012/282438. PMC 3462412. PMID 23050036.
- ^ a b c Saki M, Prakash A (June 2017). "DNA damage related crosstalk between the nucleus and mitochondria". Free Radical Biology & Medicine. 107: 216–227. doi:10.1016/j.freeradbiomed.2016.11.050. PMC 5449269. PMID 27915046.
- ^ Leon J, Sakumi K, Castillo E, Sheng Z, Oka S, Nakabeppu Y (February 2016). "8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions". Scientific Reports. 6: 22086. Bibcode:2016NatSR...622086L. doi:10.1038/srep22086. PMC 4766534. PMID 26912170.
- ^ Dahal S, Dubey S, Raghavan SC (May 2018). "Homologous recombination-mediated repair of DNA double-strand breaks operates in mammalian mitochondria". Cellular and Molecular Life Sciences. 75 (9): 1641–1655. doi:10.1007/s00018-017-2702-y. PMC 11105789. PMID 29116362. S2CID 4779650.
- ^ Odahara M, Inouye T, Fujita T, Hasebe M, Sekine Y (February 2007). "Involvement of mitochondrial-targeted RecA in the repair of mitochondrial DNA in the moss, Physcomitrella patens". Genes & Genetic Systems. 82 (1): 43–51. doi:10.1266/ggs.82.43. PMID 17396019.
- ^ Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC (January 2016). "Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions". Molecular Biology of the Cell. 27 (2): 223–235. doi:10.1091/mbc.E15-05-0260. PMC 4713127. PMID 26609070.
- ^ a b Henriquez FL, Richards TA, Roberts F, McLeod R, Roberts CW (February 2005). "The unusual mitochondrial compartment of Cryptosporidium parvum". Trends in Parasitology. 21 (2): 68–74. doi:10.1016/j.pt.2004.11.010. PMID 15664529.
- ^ John U, Lu Y, Wohlrab S, Groth M, Janouškovec J, Kohli GS, et al. (April 2019). "An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome". Science Advances. 5 (4): eaav1110. Bibcode:2019SciA....5.1110J. doi:10.1126/sciadv.aav1110. PMC 6482013. PMID 31032404.
- ^ "Veritable powerhouse – even without DNA: Parasitic algae from the dinoflagellate lineage have organized their genetic material in an unprecedented way". ScienceDaily. Archived from the original on June 24, 2019. Retrieved May 8, 2019.
- ^ a b c Zeviani M, Di Donato S (October 2004). "Mitochondrial disorders". Brain. 127 (Pt 10): 2153–2172. doi:10.1093/brain/awh259. PMID 15358637.
- ^ Taylor RW, Turnbull DM (May 2005). "Mitochondrial DNA mutations in human disease". Nature Reviews. Genetics. 6 (5): 389–402. doi:10.1038/nrg1606. PMC 1762815. PMID 15861210.
- ^ Goldman A, Khiste S, Freinkman E, Dhawan A, Majumder B, Mondal J, et al. (August 2019). "Targeting tumor phenotypic plasticity and metabolic remodeling in adaptive cross-drug tolerance". Science Signaling. 12 (595). doi:10.1126/scisignal.aas8779. PMC 7261372. PMID 31431543.
- ^ Saha T, Dash C, Jayabalan R, Khiste S, Kulkarni A, Kurmi K, et al. (January 2022). "Intercellular nanotubes mediate mitochondrial trafficking between cancer and immune cells". Nature Nanotechnology. 17 (1): 98–106. Bibcode:2022NatNa..17...98S. doi:10.1038/s41565-021-01000-4. PMC 10071558. PMID 34795441. S2CID 244349825.
- ^ Chinnery PF, Schon EA (September 2003). "Mitochondria". Journal of Neurology, Neurosurgery, and Psychiatry. 74 (9): 1188–1199. doi:10.1136/jnnp.74.9.1188. PMC 1738655. PMID 12933917.
- ^ Sherer TB, Betarbet R, Greenamyre JT (June 2002). "Environment, mitochondria, and Parkinson's disease". The Neuroscientist. 8 (3): 192–197. doi:10.1177/1073858402008003004. PMID 12061498. S2CID 24318009.
- ^ Gomez C, Bandez MJ, Navarro A (January 2007). "Pesticides and impairment of mitochondrial function in relation with the parkinsonian syndrome". Frontiers in Bioscience. 12: 1079–1093. doi:10.2741/2128. PMID 17127363.
- ^ Lim YA, Rhein V, Baysang G, Meier F, Poljak A, Raftery MJ, et al. (April 2010). "Abeta and human amylin share a common toxicity pathway via mitochondrial dysfunction". Proteomics. 10 (8): 1621–1633. doi:10.1002/pmic.200900651. PMID 20186753. S2CID 33077667.
- ^ King JV, Liang WG, Scherpelz KP, Schilling AB, Meredith SC, Tang WJ (July 2014). "Molecular basis of substrate recognition and degradation by human presequence protease". Structure. 22 (7): 996–1007. doi:10.1016/j.str.2014.05.003. PMC 4128088. PMID 24931469.
- ^ Schapira AH (July 2006). "Mitochondrial disease". Lancet. 368 (9529): 70–82. doi:10.1016/S0140-6736(06)68970-8. PMID 16815381. S2CID 27039617.
- ^ Pieczenik SR, Neustadt J (August 2007). "Mitochondrial dysfunction and molecular pathways of disease". Experimental and Molecular Pathology. 83 (1): 84–92. doi:10.1016/j.yexmp.2006.09.008. PMID 17239370.
- ^ Bugger H, Abel ED (November 2010). "Mitochondria in the diabetic heart". Cardiovascular Research. 88 (2): 229–240. doi:10.1093/cvr/cvq239. PMC 2952534. PMID 20639213.
- ^ Podolak A, Woclawek-Potocka I, Lukaszuk K (February 2022). "The Role of Mitochondria in Human Fertility and Early Embryo Development: What Can We Learn for Clinical Application of Assessing and Improving Mitochondrial DNA?". Cells. 11 (5): 797. doi:10.3390/cells11050797. PMC 8909547. PMID 35269419.
- ^ May-Panloup P, Boguenet M, Hachem HE, Bouet PE, Reynier P (January 2021). "Embryo and Its Mitochondria". Antioxidants. 10 (2): 139. doi:10.3390/antiox10020139. PMC 7908991. PMID 33498182.
- ^ Committee on the Ethical and Social Policy Considerations of Novel Techniques for Prevention of Maternal Transmission of Mitochondrial DNA Diseases; Board on Health Sciences Policy; Institute of Medicine; National Academies of Sciences, Engineering, and Medicine (March 17, 2016), Claiborne A, English R, Kahn J (eds.), "Introduction", Mitochondrial Replacement Techniques: Ethical, Social, and Policy Considerations, National Academies Press (US), retrieved December 5, 2023
- ^ Richter C, Park JW, Ames BN (September 1988). "Normal oxidative damage to mitochondrial and nuclear DNA is extensive". Proceedings of the National Academy of Sciences of the United States of America. 85 (17): 6465–6467. Bibcode:1988PNAS...85.6465R. doi:10.1073/pnas.85.17.6465. PMC 281993. PMID 3413108.
- ^ Harman D (July 1956). "Aging: a theory based on free radical and radiation chemistry". Journal of Gerontology. 11 (3): 298–300. CiteSeerX 10.1.1.663.3809. doi:10.1093/geronj/11.3.298. PMID 13332224.
- ^ Harman D (April 1972). "The biologic clock: the mitochondria?". Journal of the American Geriatrics Society. 20 (4): 145–147. doi:10.1111/j.1532-5415.1972.tb00787.x. PMID 5016631. S2CID 396830.
- ^ "Mitochondria and Aging". circuitblue.co. Archived from the original on September 29, 2017. Retrieved October 23, 2006.
- ^ Boffoli D, Scacco SC, Vergari R, Solarino G, Santacroce G, Papa S (April 1994). "Decline with age of the respiratory chain activity in human skeletal muscle". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1226 (1): 73–82. doi:10.1016/0925-4439(94)90061-2. PMID 8155742.
- ^ de Grey AD (2004). "Mitochondrial mutations in mammalian aging: an over-hasty about-turn?". Rejuvenation Research. 7 (3): 171–174. doi:10.1089/rej.2004.7.171. PMID 15588517.
- ^ Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, et al. (May 2006). "High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease". Nature Genetics. 38 (5): 515–517. doi:10.1038/ng1769. PMID 16604074. S2CID 13956928.
- ^ Mehta AR, Walters R, Waldron FM, Pal S, Selvaraj BT, Macleod MR, et al. (August 2019). "Targeting mitochondrial dysfunction in amyotrophic lateral sclerosis: a systematic review and meta-analysis". Brain Communications. 1 (1): fcz009. doi:10.1093/braincomms/fcz009. PMC 7056361. PMID 32133457. S2CID 201168692.
- ^ Mehta AR, Gregory JM, Dando O, Carter RN, Burr K, Nanda J, et al. (February 2021). "Mitochondrial bioenergetic deficits in C9orf72 amyotrophic lateral sclerosis motor neurons cause dysfunctional axonal homeostasis". Acta Neuropathologica. 141 (2): 257–279. doi:10.1007/s00401-020-02252-5. PMC 7847443. PMID 33398403.
- ^ Chiang JL, Shukla P, Pagidas K, Ahmed NS, Karri S, Gunn DD, et al. (November 2020). "Mitochondria in Ovarian Aging and Reproductive Longevity". Ageing Research Reviews. 63: 101168. doi:10.1016/j.arr.2020.101168. PMC 9375691. PMID 32896666. S2CID 221472721.
- ^ Kölliker A (1857). "Einige Bemerkungen über die Endigungen der Hautnerven und den Bau der Muskeln" [Some remarks about the terminations of the cutaneous nerves and the structure of muscles]. Zeitschrift für wissenschaftliche Zoologie (in German). 8: 311–325. Archived from the original on June 22, 2023. Retrieved June 22, 2023. On p. 316, Kölliker described mitochondria which he observed in fresh frog muscles: " ... sehr blasse rundliche Körnchen, welche in langen linienförmigen Zügen [...] wenn man einmal auf dieselben aufmerksam geworden ist." ( ... [they are] very faint round granules, which are embedded in the [muscle's] contractile substance in long linear trains. These granules are located in the whole thickness of the muscle fiber, on the surface as in the interior, and [they] are so numerous that they appear as a not unimportant element of the muscle fibers, once one has become alert to them.) Kölliker said (p. 321) that he had found mitochondria in the muscles of other animals. In Figure 3 of Table XIV Archived June 22, 2023, at the Wayback Machine, Kölliker depicted mitochondria in frog muscles.
- ^ a b c d e f g h i j Ernster L, Schatz G (December 1981). "Mitochondria: a historical review". The Journal of Cell Biology. 91 (3 Pt 2): 227s – 255s. doi:10.1083/jcb.91.3.227s. PMC 2112799. PMID 7033239.
- ^ Altmann R (1890). Die Elementarorganismen und ihre Beziehungen zu den Zellen [Elementary Organisms and Their Relations to Cells] (in German). Leipzig, Germany: Veit & Co. p. 125. Archived from the original on June 23, 2023. Retrieved June 23, 2023. From p. 125: "Da auch sonst mancherlei Umstände dafür sprechen, dass Mikroorganismen und Granula einander gleichwerthig sind und Elementarorganismen vorstellen, welche sich überall finden, wo lebendige Kräfte ausgelöst werden, so wollen wir sie mit dem gemeinschaftlichen Namen der Bioblasten bezeichnen." (Since otherwise some circumstances indicate that microorganisms and granula are equivalent to each other and suggest elementary organisms, which are to be found wherever living forces are unleashed, we will designate them with the collective name of "bioblasts".)
- ^ "mitochondria". Online Etymology Dictionary. Archived from the original on March 4, 2016. Retrieved May 23, 2013.
- ^ Benda C (1898). "Ueber die Spermatogenese der Vertebraten und höherer Evertebraten. II. Theil: Die Histiogenese der Spermien" [On spermatogenesis in vertebrates and higher invertebrates. Part II: The histogenesis of sperm.]. Archiv für Physiologie (in German). 1898: 393–398. Archived from the original on February 24, 2019. Retrieved January 14, 2018. From p. 397: After Brenda states that " ... ich bereits in vielen Zellarten aller möglichen Thierclassen gefunden habe, ... " ( ... I have already found [them (mitochondria)] in many types of cells of all possible classes of animals, ... ), he suggests: "Ich möchte vorläufig vorschlagen, ihnen als Mitochondria eine besondere Stellung vorzubehalten, die ich in weiteren Arbeiten begründen werde." (I would like to suggest provisionally reserving for them, as "mitochondria", a special status which I will justify in further work.)
- Benda C (1899). "Weitere Mitteilungen über die Mitochondria" [Further reports on mitochondria]. Archiv für Physiologie: Verhandlungen der Berliner Physiologischen Gesellschaft (in German). 1899: 376–383. Archived from the original on June 23, 2023. Retrieved June 23, 2023.
- ^ Michaelis L (1900). "Die vitale Farbung, eine Darstellungsmethode der Zellgranula" [The vital stain: a method of imaging cell granules]. Archiv für Mikroskopische Anatomie und Entwicklungsgeschichte [Archive for Microscopic Anatomy and Ontogenesis] (in German). 55: 558–575. doi:10.1007/BF02977747. S2CID 85010030. Archived from the original on June 23, 2023. Retrieved June 23, 2023.
- ^ Ernster's citation Meves F (May 1908). "Die Chondriosomen als Träger erblicher Anlagen. Cytologische Studien am Hühnerembryo". Archiv für Mikroskopische Anatomie. 72 (1): 816–867. doi:10.1007/BF02982402. S2CID 85047330. Archived from the original on January 9, 2020. Retrieved June 24, 2019. is wrong, correct citation is Meves F (1904). "Über das Vorkommen von Mitochondrien bezw. Chondromiten in Pflanzenzellen". Ber. Dtsch. Bot. Ges. 22: 284–286. doi:10.1111/j.1438-8677.1904.tb05237.x. S2CID 257822147., cited in Meves' 1908 paper and in Schmidt EW (1913). "Pflanzliche Mitochondrien". Progressus Rei Botanicae. 4: 164–183. Retrieved September 21, 2012., with confirmation of Nymphaea alba
- ^ Kingsbury BF (1912). "Cytoplasmic fixation". The Anatomical Record. 6 (2): 39–52. doi:10.1002/ar.1090060202. S2CID 221400422. Archived from the original on June 23, 2023. Retrieved June 23, 2023. From p. 47: " ... the mitochondria are the structural expression thereof [i.e., of the chemical reducing processes in the cytoplasm], ... "
- ^ Warburg O (1913). "Über sauerstoffatmende Körnchen aus Leberzellen und über Sauerstoffatmung in Berkefeld-Filtraten wässriger Leberextrake" [On respiring granules from liver cells and on respiration in Berkefeld filtrates of aqueous liver extracts]. Pflügers Archiv für die gesamte Physiologie des Menschen und der Tiere (Pfluger's Archive for All Physiology of Humans and Animals) (in German). 154: 599–617. Archived from the original on June 23, 2023. Retrieved June 23, 2023.
- ^ Milane L, Trivedi M, Singh A, Talekar M, Amiji M (June 2015). "Mitochondrial biology, targets, and drug delivery". Journal of Controlled Release. 207: 40–58. doi:10.1016/j.jconrel.2015.03.036. PMID 25841699.
- ^ Martin WF, Garg S, Zimorski V (September 2015). "Endosymbiotic theories for eukaryote origin". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 370 (1678): 20140330. doi:10.1098/rstb.2014.0330. PMC 4571569. PMID 26323761.
General
 This article incorporates public domain material from Science Primer. NCBI. Archived from the original on December 8, 2009.
This article incorporates public domain material from Science Primer. NCBI. Archived from the original on December 8, 2009.
External links
- Lane N (2016). The Vital Question: Energy, Evolution, and the Origins of Complex Life. WW Norton & Company. ISBN 978-0393352979.
- Powering the Cell Mitochondria Archived August 17, 2022, at the Wayback Machine – XVIVO Scientific Animation
- Mitodb.com Archived July 3, 2013, at the Wayback Machine – The mitochondrial disease database.
- Mitochondria Atlas Archived June 29, 2012, at the Wayback Machine at University of Mainz
- Mitochondria Research Portal at mitochondrial.net
- Mitochondria: Architecture dictates function at cytochemistry.net
- Mitochondria links at University of Alabama
- MIP Archived May 23, 2016, at the Portuguese Web Archive Mitochondrial Physiology Society
- 3D structures of proteins from inner mitochondrial membrane Archived September 13, 2011, at the Wayback Machine at University of Michigan
- 3D structures of proteins associated with outer mitochondrial membrane Archived September 13, 2011, at the Wayback Machine at University of Michigan
- Mitochondrial Protein Partnership at University of Wisconsin
- MitoMiner – A mitochondrial proteomics database[permanent dead link] at MRC Mitochondrial Biology Unit
- Mitochondrion – Cell Centered Database Archived October 7, 2011, at the Wayback Machine
- Mitochondrion Reconstructed by Electron Tomography Archived November 14, 2012, at the Wayback Machine at San Diego State University
- Video Clip of Rat-liver Mitochondrion from Cryo-electron Tomography






