ALDH2
| ALDH2 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ALDH2, ALDH-E2, ALDHI, ALDM, aldehyde dehydrogenase 2 family (mitochondrial), aldehyde dehydrogenase 2 family member | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 100650; MGI: 99600; HomoloGene: 55480; GeneCards: ALDH2; OMA:ALDH2 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Aldehyde dehydrogenase, mitochondrial is an enzyme that in humans is encoded by the ALDH2 gene located on chromosome 12.[5][6] ALDH2 belongs to the aldehyde dehydrogenase family of enzymes. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. ALDH2 has a low Km for acetaldehyde, and is localized in mitochondrial matrix. The other liver isozyme, ALDH1, localizes to the cytosol.[7]
Most White people have both major isozymes, while approximately 36% of East Asians have the cytosolic isozyme but not a functional mitochondrial isozyme. A remarkably higher frequency of acute alcohol intoxication among East Asians than among Whites could be related to this absence of a catalytically active form of ALDH2. The increased exposure to acetaldehyde in individuals with the catalytically inactive form may also confer greater susceptibility to many types of cancer.[8]
Gene
[edit]The ALDH2 gene is about 44 kbp in length and contains at least 13 exons which encode 517 amino acid residues. Except for the signal NH2-terminal peptide, which is absent in the mature enzyme, the amino acid sequence deduced from the exons coincided with the reported primary structure of human liver ALDH2. Several introns contain Alu repetitive sequences. A TATA-like sequence (TTATAAAA) and a CAAT-like sequence (GTCATCAT) are located 473 and 515 bp, respectively, upstream from the translation initiation codon.[9]
Enzyme structure
[edit]ALDH2 is a tetrameric enzyme that contains three domains; two dinucleotide-binding domains and a three-stranded beta-sheet domain. The active site of ALDH2 is divided into two halves by the nicotinamide ring of nicotinamide adenine dinucleotide (NAD+). Adjacent to the A-side (Pro-R) of the nicotinamide ring is a cluster of three cysteines (Cys301, Cys302 and Cys303) and adjacent to the B-side (Pro-S) are Thr244, Glu268, Glu476 and an ordered water molecule bound to Thr244 and Glu476.[10] Although there is a recognizable Rossmann fold, the coenzyme-binding region of ALDH2 binds NAD+ in a manner not seen in other NAD+-binding enzymes. The positions of the residues near the nicotinamide ring of NAD+ suggest a chemical mechanism whereby Glu268 functions as a general base through a bound water molecule. The sidechain amide nitrogen of Asn169 and the peptide nitrogen of Cys302 are in position to stabilize the oxyanion present in the tetrahedral transition state prior to hydride transfer. The functional importance of residue Glu487 now appears to be due to indirect interactions of this residue with the substrate-binding site via Arg264 and Arg475.[11]
Function
[edit]Mitochondrial aldehyde dehydrogenase belongs to the aldehyde dehydrogenase family of enzymes that catalyze the chemical transformation from acetaldehyde to acetic acid. Aldehyde dehydrogenase is the second enzyme of the major oxidative pathway of alcohol metabolism. Human ALDH2 is especially efficient on acetaldehyde compared to ALDH1.[12]
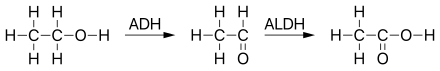
Additionally, ALDH2 functions as a protector against oxidative stress.[13]
Genetic variation
[edit]| SNP: ALDH2*2 | |
|---|---|
| Name(s) | g.42421G>A, Glu504Lys |
| Gene | ALDH2 |
| Chromosome | 12 |
| Region | Exon |
| External databases | |
| Ensembl | Human SNPView |
| dbSNP | 671 |
| HapMap | 671 |
| SNPedia | 671 |
The inactivating ALDH2*2 mutation is "the most common single point mutation in humans".[14] This mutation is found in very few White people, but about 50% of East Asians are heterozygous for this mutation. The ALDH2*2 allele encodes lysine instead of glutamic acid at amino acid 487,[15] distorting the NAD+ binding site.[16][17] ALDH2 assembles and functions as a tetramer and requires all four of its components to be active in order to metabolize acetaldehyde. People heterozygous for ALDH2*2 have only 10% to 45% enzyme activity, while those homozygous for ALDH2*2 have as little as 1% to 5% remaining activity.[18]
The lack of ALDH2 activity has a number of consequences, detailed in section § Inhibition and genetic deficiency below.
Distribution
[edit]In the overall Japanese population, about 57% of individuals are homozygous for the normal allele, 40% are heterozygous for the ALDH2*2 allele, and 3% are homozygous for the ALDH2*2 allele.[15]
Clinical significance
[edit]Inhibition and genetic deficiency
[edit]Alcohol metabolism
[edit]The best-known consequence of ALDH2 dysfunction is in relation to the consumption of ethanol. People heterozygous or homozygous for the ALDH2*2 metabolize ethanol to acetaldehyde normally but metabolize acetaldehyde poorly. As a result, they accumulate increased levels of acetaldehyde after consumption of alcoholic beverages. Effects include facial flushing (i.e. the "Alcohol flush reaction"), urticaria, systemic dermatitis, and alcohol-induced respiratory reactions such as rhinitis and the exacerbation of asthma bronchoconstriction.[19] The cited allergic reaction-like symptoms: (a) do not appear due to classical IgE or T cell-related allergen-induced reactions but rather the actions of acetaldehyde in stimulating the release of histamine, a probable mediating cause of these symptoms; (b) typically occur within 30–60 minutes of ingesting alcoholic beverages; and (c) occur in other Asian as well as non-Asian individuals that are either seriously defective in metabolizing ingested ethanol past acetaldehyde to acetic acid or, alternatively, that metabolize ethanol too rapidly for ALDH2 processing.[19][20]
People with a genetic ALDH2*2 deficiency have historically had a lower likelihood of developing alcoholism, both from stronger adverse effects and a possible reduction of dopamine release.[21] However, this effect is not absolute: during the 1980s, there has been a steady increase in the number of Japanese alcoholics who carry the ALDH2*2 mutation. A strong social pressure to drink have overcome this genetic barrier to alcoholism.[22] Disulfiram, which inhibits ALDH2 and causes a similar effect, has been used as an alcohol-quitting aid.[21]
Various conditions
[edit]More recently, ALDH2 has been implicated in a number of pathways beyond alcohol metabolism. ALDH2 dysfunction is supposedly associated with a variety of human diseases including diabetes, neurodegenerative diseases, cardiovascular diseases and stroke, cancer, Fanconi anemia, pain, osteoporosis, and the process of aging.[14] The inactivating ALDH2 rs671 polymorphism, present in up to 8% of the global population and in up to 50% of the East Asian population, is associated with increased risk of cardiovascular conditions such as coronary artery disease, alcohol-induced cardiac dysfunction, pulmonary arterial hypertension, heart failure and drug-induced cardiotoxicity.[23]
Alzheimer's disease
[edit]A case-control study in a Japanese population showed that deficiency of ALDH2 activity influences the risk for late-onset Alzheimer's disease.[13] The ALDH2 knockout mice display age-related memory deficits in various tasks, as well as endothelial dysfunction, brain atrophy, and other Alzheimer's disease-associated pathologies, including marked increases in lipid peroxidation products, amyloid-beta, p-tau and activated caspases. These behavioral and biochemical Alzheimer's disease-like deficits were efficiently ameliorated when these mice were treated with isotope-reinforced lipids (deuterated polyunsaturated fatty acids).[24]
Activation
[edit]An activator of ALDH2 enzymatic activity, Alda-1 (N-(1,3-benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide), has been shown to reduce ischemia-induced cardiac damage caused by myocardial infarction.[25]
Interactions
[edit]ALDH2 has been shown to interact with GroEL.[26]
See also
[edit]- Acetaldehyde dehydrogenase
- Alcohol dehydrogenase
- Alcohol flush reaction
- Alcohol-induced respiratory reactions
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000111275 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000029455 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Yoshida A, Ikawa M, Hsu LC, Tani K (1985). "Molecular abnormality and cDNA cloning of human aldehyde dehydrogenases". Alcohol. 2 (1): 103–6. doi:10.1016/0741-8329(85)90024-2. PMID 4015823.
- ^ Hsu LC, Tani K, Fujiyoshi T, Kurachi K, Yoshida A (Jun 1985). "Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2". Proceedings of the National Academy of Sciences of the United States of America. 82 (11): 3771–5. Bibcode:1985PNAS...82.3771H. doi:10.1073/pnas.82.11.3771. PMC 397869. PMID 2987944.
- ^ "Entrez Gene: ALDH2 aldehyde dehydrogenase 2 family (mitochondrial)".
- ^ Seitz HK, Meier P (2007). "The role of acetaldehyde in upper digestive tract cancer in alcoholics". Transl Res. 149 (6): 293–7. doi:10.1016/j.trsl.2006.12.002. PMID 17543846.
- ^ Hsu LC, Bendel RE, Yoshida A (Jan 1988). "Genomic structure of the human mitochondrial aldehyde dehydrogenase gene". Genomics. 2 (1): 57–65. doi:10.1016/0888-7543(88)90109-7. PMID 2838413.
- ^ González-Segura L, Ho KK, Perez-Miller S, Weiner H, Hurley TD (Feb 2013). "Catalytic contribution of threonine 244 in human ALDH2". Chemico-Biological Interactions. 202 (1–3): 32–40. Bibcode:2013CBI...202...32G. doi:10.1016/j.cbi.2012.12.009. PMC 3602351. PMID 23295226.
- ^ Steinmetz CG, Xie P, Weiner H, Hurley TD (May 1997). "Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion". Structure. 5 (5): 701–11. doi:10.1016/s0969-2126(97)00224-4. PMID 9195888.
- ^ Wang MF, Han CL, Yin SJ (16 March 2009). "Substrate specificity of human and yeast aldehyde dehydrogenases". Chemico-Biological Interactions. 178 (1–3): 36–9. Bibcode:2009CBI...178...36W. doi:10.1016/j.cbi.2008.10.002. PMID 18983993.
- ^ a b Ohta S, Ohsawa I, Kamino K, Ando F, Shimokata H (Apr 2004). "Mitochondrial ALDH2 deficiency as an oxidative stress". Annals of the New York Academy of Sciences. 1011 (1): 36–44. Bibcode:2004NYASA1011...36O. doi:10.1196/annals.1293.004. PMID 15126281. S2CID 28571902.
- ^ a b Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D (2014). "Targeting Aldehyde Dehydrogenase 2: New Therapeutic Opportunities". Physiological Reviews. 94 (1): 1–34. doi:10.1152/physrev.00017.2013. PMC 3929114. PMID 24382882.
- ^ a b Takao A, Shimoda T, Kohno S, Asai S, Harda S (May 1998). "Correlation between alcohol-induced asthma and acetaldehyde dehydrogenase-2 genotype". The Journal of Allergy and Clinical Immunology. 101 (5): 576–80. doi:10.1016/S0091-6749(98)70162-9. PMID 9600491.
- ^ Larson HN, Weiner H, Hurley TD (August 2005). "Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase "Asian" variant". The Journal of Biological Chemistry. 280 (34): 30550–6. doi:10.1074/jbc.M502345200. PMC 1262676. PMID 15983043.
- ^ Chang HY, Mitchell A (May 2014). "Dionysian Mysteries The Aldehyde Dehydrogenase (aldh) Family". InterPro Protein Focus.
- ^ Ma C, Yu B, Zhang W, Wang W, Zhang L, Zeng Q (11 September 2017). "Associations between aldehyde dehydrogenase 2 (ALDH2) rs671 genetic polymorphisms, lifestyles and hypertension risk in Chinese Han people". Scientific Reports. 7 (1): 11136. Bibcode:2017NatSR...711136M. doi:10.1038/s41598-017-11071-w. PMC 5593832. PMID 28894224.
- ^ a b Adams KE, Rans TS (Dec 2013). "Adverse reactions to alcohol and alcoholic beverages". Annals of Allergy, Asthma & Immunology. 111 (6): 439–45. doi:10.1016/j.anai.2013.09.016. PMID 24267355.
- ^ Linneberg A, Gonzalez-Quintela A, Vidal C, Jørgensen T, Fenger M, Hansen T, Pedersen O, Husemoen LL (Jan 2010). "Genetic determinants of both ethanol and acetaldehyde metabolism influence alcohol hypersensitivity and drinking behaviour among Scandinavians". Clinical and Experimental Allergy. 40 (1): 123–30. doi:10.1111/j.1365-2222.2009.03398.x. PMID 20205700. S2CID 40246805.
- ^ a b Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D (2014). "Targeting Aldehyde Dehydrogenase 2: New Therapeutic Opportunities". Physiological Reviews. 94 (1): 1–34. doi:10.1152/physrev.00017.2013. PMC 3929114. PMID 24382882.
- ^ Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H (March 1994). "Aldehyde dehydrogenase genotypes in Japanese alcoholics". Lancet. 343 (8899): 741–2. doi:10.1016/S0140-6736(94)91629-2. PMID 7907720. S2CID 41404745.
- ^ Zhang J, Guo Y, Zhao X, Pang J, Pan C, Wang J, Wei S, Yu X, Zhang C, Chen Y, Yin H, Xu F (2023). "The role of aldehyde dehydrogenase 2 in cardiovascular disease". Nature Reviews. Cardiology. 20 (7): 495–509. doi:10.1038/s41569-023-00839-5. PMID 36781974. S2CID 256845243.
- ^ Elharram A, Czegledy NM, Golod M, Milne GL, Pollock E, Bennett BM, Shchepinov MS (December 2017). "Deuterium-reinforced polyunsaturated fatty acids improve cognition in a mouse model of sporadic Alzheimer's disease". The FEBS Journal. 284 (23): 4083–4095. doi:10.1111/febs.14291. PMC 5716852. PMID 29024570.
- ^ Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D (Sep 2008). "Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart". Science. 321 (5895): 1493–5. Bibcode:2008Sci...321.1493C. doi:10.1126/science.1158554. PMC 2741612. PMID 18787169.
- ^ Lee KH, Kim HS, Jeong HS, Lee YS (Oct 2002). "Chaperonin GroESL mediates the protein folding of human liver mitochondrial aldehyde dehydrogenase in Escherichia coli". Biochemical and Biophysical Research Communications. 298 (2): 216–24. doi:10.1016/S0006-291X(02)02423-3. PMID 12387818.
Further reading
[edit]- Yoshida A (1992). "Molecular genetics of human aldehyde dehydrogenase". Pharmacogenetics. 2 (4): 139–47. doi:10.1097/00008571-199208000-00001. PMID 1306115.
- Chao YC, Liou SR, Tsai SF, Yin SJ (1993). "Dominance of the mutant ALDH2(2) allele in the expression of human stomach aldehyde dehydrogenase-2 activity". Proc. Natl. Sci. Counc. Repub. China B. 17 (3): 98–102. PMID 8290656.
- Crabb DW, Edenberg HJ, Bosron WF, Li TK (1989). "Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant". J. Clin. Invest. 83 (1): 314–6. doi:10.1172/JCI113875. PMC 303676. PMID 2562960.
- Hsu LC, Bendel RE, Yoshida A (1988). "Genomic structure of the human mitochondrial aldehyde dehydrogenase gene". Genomics. 2 (1): 57–65. doi:10.1016/0888-7543(88)90109-7. PMID 2838413.
- Hsu LC, Tani K, Fujiyoshi T, Kurachi K, Yoshida A (1985). "Cloning of cDNAs for human aldehyde dehydrogenases 1 and 2". Proc. Natl. Acad. Sci. U.S.A. 82 (11): 3771–5. Bibcode:1985PNAS...82.3771H. doi:10.1073/pnas.82.11.3771. PMC 397869. PMID 2987944.
- Braun T, Grzeschik KH, Bober E, Singh S, Agarwal DP, Goedde HW (1986). "The structural gene for the mitochondrial aldehyde dehydrogenase maps to human chromosome 12". Hum. Genet. 73 (4): 365–7. doi:10.1007/BF00279102. PMID 3017845. S2CID 28795641.
- Braun T, Bober E, Singh S, Agarwal DP, Goedde HW (1987). "Isolation and sequence analysis of a full length cDNA clone coding for human mitochondrial aldehyde dehydrogenase". Nucleic Acids Res. 15 (7): 3179. doi:10.1093/nar/15.7.3179. PMC 340920. PMID 3562250.
- Braun T, Bober E, Singh S, Agarwal DP, Goedde HW (1987). "Evidence for a signal peptide at the amino-terminal end of human mitochondrial aldehyde dehydrogenase". FEBS Lett. 215 (2): 233–6. Bibcode:1987FEBSL.215..233B. doi:10.1016/0014-5793(87)80152-7. PMID 3582651. S2CID 42627599.
- Agarwal DP, Goedde HW (1987). "Human aldehyde dehydrogenase isozymes and alcohol sensitivity". Isozymes Curr. Top. Biol. Med. Res. 16: 21–48. PMID 3610592.
- Hempel J, Höög JO, Jörnvall H (1987). "Mitochondrial aldehyde dehydrogenase. Homology of putative targeting sequence to that of carbamyl phosphate synthetase I revealed by correlation of cDNA and protein data". FEBS Lett. 222 (1): 95–8. Bibcode:1987FEBSL.222...95H. doi:10.1016/0014-5793(87)80198-9. PMID 3653404. S2CID 33980993.
- Yoshida A, Ikawa M, Hsu LC, Tani K (1985). "Molecular abnormality and cDNA cloning of human aldehyde dehydrogenases". Alcohol. 2 (1): 103–6. doi:10.1016/0741-8329(85)90024-2. PMID 4015823.
- Hempel J, Kaiser R, Jörnvall H (1985). "Mitochondrial aldehyde dehydrogenase from human liver. Primary structure, differences in relation to the cytosolic enzyme, and functional correlations". Eur. J. Biochem. 153 (1): 13–28. doi:10.1111/j.1432-1033.1985.tb09260.x. PMID 4065146.
- Yoshida A, Huang IY, Ikawa M (1984). "Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals". Proc. Natl. Acad. Sci. U.S.A. 81 (1): 258–61. Bibcode:1984PNAS...81..258Y. doi:10.1073/pnas.81.1.258. PMC 344651. PMID 6582480.
- Xiao Q, Weiner H, Johnston T, Crabb DW (1995). "The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced HeLa cells". J. Clin. Invest. 96 (5): 2180–6. doi:10.1172/JCI118272. PMC 185867. PMID 7593603.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Novoradovsky A, Tsai SJ, Goldfarb L, Peterson R, Long JC, Goldman D (1995). "Mitochondrial aldehyde dehydrogenase polymorphism in Asian and American Indian populations: detection of new ALDH2 alleles". Alcohol. Clin. Exp. Res. 19 (5): 1105–10. doi:10.1111/j.1530-0277.1995.tb01587.x. PMID 8561277.
- Xiao Q, Weiner H, Crabb DW (1996). "The mutation in the mitochondrial aldehyde dehydrogenase (ALDH2) gene responsible for alcohol-induced flushing increases turnover of the enzyme tetramers in a dominant fashion". J. Clin. Invest. 98 (9): 2027–32. doi:10.1172/JCI119007. PMC 507646. PMID 8903321.
External links
[edit]- ALDH2 protein, human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Human ALDH2 genome location and ALDH2 gene details page in the UCSC Genome Browser.