RNA virus
Parts of this article (those related to taxonomy in baltimore sections [ICTV release 2018b→2019]) need to be updated. (January 2021) |

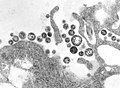
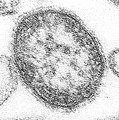
An RNA virus is a virus characterized by a ribonucleic acid (RNA) based genome.[1] The genome can be single-stranded RNA (ssRNA) or double-stranded (dsRNA).[2] Notable human diseases caused by RNA viruses include influenza, SARS, MERS, COVID-19, Dengue virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio, mumps, and measles.
All known RNA viruses, that is viruses that use a homologous RNA-dependent polymerase for replication, are categorized by the International Committee on Taxonomy of Viruses (ICTV) into the realm Riboviria.[3] This includes RNA viruses belonging to Group III, Group IV or Group V of the Baltimore classification system as well as Group VI. Group VI viruses are retroviruses, viruses with RNA genetic material that use DNA intermediates in their life cycle including HIV-1 and HIV-2 which cause AIDS.
The majority of such RNA viruses fall into the kingdom Orthornavirae and the rest have a positioning not yet defined.[4] The realm does not contain all RNA viruses: Deltavirus, Avsunviroidae, and Pospiviroidae are taxa of RNA viruses that were mistakenly included in 2019,[a] but corrected in 2020.[5]
Characteristics
[edit]Single-stranded RNA viruses and RNA Sense
[edit]RNA viruses can be further classified according to the sense or polarity of their RNA into negative-sense and positive-sense, or ambisense RNA viruses. Positive-sense viral RNA is similar to mRNA and thus can be immediately translated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA-dependent RNA polymerase before translation. Purified RNA of a positive-sense virus can directly cause infection though it may be less infectious than the whole virus particle. In contrast, purified RNA of a negative-sense virus is not infectious by itself as it needs to be transcribed into positive-sense RNA; each virion can be transcribed to several positive-sense RNAs. Ambisense RNA viruses resemble negative-sense RNA viruses, except they translate genes from their negative and positive strands.[6]
Double-stranded RNA viruses
[edit]
The double-stranded (ds)RNA viruses represent a diverse group of viruses that vary widely in host range (humans, animals, plants, fungi,[b] and bacteria), genome segment number (one to twelve), and virion organization (Triangulation number, capsid layers, spikes, turrets, etc.). Members of this group include the rotaviruses, which are the most common cause of gastroenteritis in young children, and picobirnaviruses, which are the most common virus in fecal samples of both humans and animals with or without signs of diarrhea. Bluetongue virus is an economically important pathogen that infects cattle and sheep. In recent years, progress has been made in determining atomic and subnanometer resolution structures of a number of key viral proteins and virion capsids of several dsRNA viruses, highlighting the significant parallels in the structure and replicative processes of many of these viruses.[2][page needed]
Mutation rates
[edit]RNA viruses generally have very high mutation rates compared to DNA viruses,[8] because viral RNA polymerases lack the proofreading ability of DNA polymerases.[9] The genetic diversity of RNA viruses is one reason why it is difficult to make effective vaccines against them.[10] Retroviruses also have a high mutation rate even though their DNA intermediate integrates into the host genome (and is thus subject to host DNA proofreading once integrated), because errors during reverse transcription are embedded into both strands of DNA before integration.[11] Some genes of RNA virus are important to the viral replication cycles and mutations are not tolerated. For example, the region of the hepatitis C virus genome that encodes the core protein is highly conserved,[12] because it contains an RNA structure involved in an internal ribosome entry site.[13]
Sequence complexity
[edit]On average, dsRNA viruses show a lower sequence redundancy relative to ssRNA viruses. Contrarily, dsDNA viruses contain the most redundant genome sequences while ssDNA viruses have the least.[14] The sequence complexity of viruses has been shown to be a key characteristic for accurate reference-free viral classification.[14]
Replication
[edit]There are three distinct groups of RNA viruses depending on their genome and mode of replication:
- Double-stranded RNA viruses (Group III) contain from one to a dozen different RNA molecules, each coding for one or more viral proteins.
- Positive-sense ssRNA viruses (Group IV) have their genome directly utilized as mRNA, with host ribosomes translating it into a single protein that is modified by host and viral proteins to form the various proteins needed for replication. One of these includes RNA-dependent RNA polymerase (RNA replicase), which copies the viral RNA to form a double-stranded replicative form. In turn, this dsRNA directs the formation of new viral RNA.
- Negative-sense ssRNA viruses (Group V) must have their genome copied by an RNA replicase to form positive-sense RNA. This means that the virus must bring along with it the enzyme RNA replicase. The positive-sense RNA molecule then acts as viral mRNA, which is translated into proteins by the host ribosomes.
Retroviruses (Group VI) have a single-stranded RNA genome but, in general, are not considered RNA viruses because they use DNA intermediates to replicate. Reverse transcriptase, a viral enzyme that comes from the virus itself after it is uncoated, converts the viral RNA into a complementary strand of DNA, which is copied to produce a double-stranded molecule of viral DNA. After this DNA is integrated into the host genome using the viral enzyme integrase, expression of the encoded genes may lead to the formation of new virions.
Recombination
[edit]Numerous RNA viruses are capable of genetic recombination when at least two viral genomes are present in the same host cell.[15] Very rarely viral RNA can recombine with host RNA.[16] RNA recombination appears to be a major driving force in determining genome architecture and the course of viral evolution among Picornaviridae ((+)ssRNA), e.g. poliovirus.[17] In the Retroviridae ((+)ssRNA), e.g. HIV, damage in the RNA genome appears to be avoided during reverse transcription by strand switching, a form of recombination.[18][19][20] Recombination also occurs in the Reoviridae (dsRNA), e.g. reovirus; Orthomyxoviridae ((-)ssRNA), e.g. influenza virus;[20] and Coronaviridae ((+)ssRNA), e.g. SARS.[21] Recombination in RNA viruses appears to be an adaptation for coping with genome damage.[15] Recombination can occur infrequently between animal viruses of the same species but of divergent lineages. The resulting recombinant viruses may sometimes cause an outbreak of infection in humans.[21]
Classification
[edit]This section may require cleanup to meet Wikipedia's quality standards. The specific problem is: outdated and redundant with riboviria article as well as later text; see DNA virus for a clean integration between ICTV higher-order and Baltimore. (January 2021) |
Classification is based principally on the type of genome (double-stranded, negative- or positive-single-strand) and gene number and organization. Currently, there are 5 orders and 47 families of RNA viruses recognized. There are also many unassigned species and genera.
Related to but distinct from the RNA viruses are the viroids and the RNA satellite viruses. These are not currently classified as RNA viruses and are described on their own pages.
A study of several thousand RNA viruses has shown the presence of at least five main taxa: a levivirus and relatives group; a picornavirus supergroup; an alphavirus supergroup plus a flavivirus supergroup; the dsRNA viruses; and the -ve strand viruses.[22] The lentivirus group appears to be basal to all the remaining RNA viruses. The next major division lies between the picornasupragroup and the remaining viruses. The dsRNA viruses appear to have evolved from a +ve RNA ancestor and the -ve RNA viruses from within the dsRNA viruses. The closest relation to the -ve stranded RNA viruses is the Reoviridae.
This is the single largest group of RNA viruses[23] and has been organized by the ICTV into the phyla Kitrinoviricota, Lenarviricota, and Pisuviricota in the kingdom Orthornavirae and realm Riboviria.[24]
Positive-strand RNA viruses can also be classified based on the RNA-dependent RNA polymerase. Three groups have been recognised:[25]
- Bymoviruses, comoviruses, nepoviruses, nodaviruses, picornaviruses, potyviruses, sobemoviruses and a subset of luteoviruses (beet western yellows virus and potato leafroll virus)—the picorna like group (Picornavirata).
- Carmoviruses, dianthoviruses, flaviviruses, pestiviruses, statoviruses, tombusviruses, single-stranded RNA bacteriophages, hepatitis C virus and a subset of luteoviruses (barley yellow dwarf virus)—the flavi like group (Flavivirata).
- Alphaviruses, carlaviruses, furoviruses, hordeiviruses, potexviruses, rubiviruses, tobraviruses, tricornaviruses, tymoviruses, apple chlorotic leaf spot virus, beet yellows virus and hepatitis E virus—the alpha like group (Rubivirata).
A division of the alpha-like (Sindbis-like) supergroup on the basis of a novel domain located near the N termini of the proteins involved in viral replication has been proposed.[26] The two groups proposed are: the 'altovirus' group (alphaviruses, furoviruses, hepatitis E virus, hordeiviruses, tobamoviruses, tobraviruses, tricornaviruses and probably rubiviruses); and the 'typovirus' group (apple chlorotic leaf spot virus, carlaviruses, potexviruses and tymoviruses).
The alpha like supergroup can be further divided into three clades: the rubi-like, tobamo-like, and tymo-like viruses.[27]
Additional work has identified five groups of positive-stranded RNA viruses containing four, three, three, three, and one order(s), respectively.[28] These fourteen orders contain 31 virus families (including 17 families of plant viruses) and 48 genera (including 30 genera of plant viruses). This analysis suggests that alphaviruses and flaviviruses can be separated into two families—the Togaviridae and Flaviridae, respectively—but suggests that other taxonomic assignments, such as the pestiviruses, hepatitis C virus, rubiviruses, hepatitis E virus, and arteriviruses, may be incorrect. The coronaviruses and toroviruses appear to be distinct families in distinct orders and not distinct genera of the same family as currently classified. The luteoviruses appear to be two families rather than one, and apple chlorotic leaf spot virus appears not to be a closterovirus but a new genus of the Potexviridae.
Evolution
[edit]The evolution of the picornaviruses based on an analysis of their RNA polymerases and helicases appears to date to the divergence of eukaryotes.[29] Their putative ancestors include the bacterial group II retroelements, the family of HtrA proteases and DNA bacteriophages.
Partitiviruses are related to and may have evolved from a totivirus ancestor.[30]
Hypoviruses and barnaviruses appear to share an ancestry with the potyvirus and sobemovirus lineages respectively.[30]
Double-stranded RNA viruses
[edit]This analysis also suggests that the dsRNA viruses are not closely related to each other but instead belong to four additional classes—Birnaviridae, Cystoviridae, Partitiviridae, and Reoviridae—and one additional order (Totiviridae) of one of the classes of positive ssRNA viruses in the same subphylum as the positive-strand RNA viruses.
One study has suggested that there are two large clades: One includes the families Caliciviridae, Flaviviridae, and Picornaviridae and a second that includes the families Alphatetraviridae, Birnaviridae, Cystoviridae, Nodaviridae, and Permutotretraviridae.[31]
Negative strand RNA viruses
[edit]These viruses have multiple types of genome ranging from a single RNA molecule up to eight segments. Despite their diversity it appears that they may have originated in arthropods and to have diversified from there.[32]
Satellite viruses
[edit]A number of satellite viruses—viruses that require the assistance of another virus to complete their life cycle—are also known. Their taxonomy has yet to be settled. The following four genera have been proposed for positive sense single stranded RNA satellite viruses that infect plants—Albetovirus, Aumaivirus, Papanivirus and Virtovirus.[33] A family—Sarthroviridae which includes the genus Macronovirus—has been proposed for the positive sense single stranded RNA satellite viruses that infect arthropods.
Group III – dsRNA viruses
[edit]There are twelve families and a number of unassigned genera and species recognised in this group.[9]
- Family Amalgaviridae
- Family Birnaviridae
- Family Chrysoviridae
- Family Cystoviridae
- Family Endornaviridae
- Family Hypoviridae
- Family Megabirnaviridae
- Family Partitiviridae
- Family Picobirnaviridae
- Family Reoviridae – includes Rotavirus
- Family Totiviridae
- Family Quadriviridae
- Genus Botybirnavirus
- Unassigned species
Group IV – positive-sense ssRNA viruses
[edit]There are three orders and 34 families recognised in this group. In addition, there are a number of unclassified species and genera.
- Order Nidovirales
- Family Arteriviridae
- Family Coronaviridae – includes Human coronavirus (common cold viruses HCoV-229E, HCoV-HKU1, HCoV-NL63, and HCoV-OC43), MERS-CoV, SARS-CoV-1 and SARS-CoV-2
- Family Mesoniviridae
- Family Roniviridae
- Order Picornavirales
- Family Dicistroviridae
- Family Iflaviridae
- Family Marnaviridae
- Family Picornaviridae – includes Poliovirus, Rhinovirus (a common cold virus), Hepatitis A virus
- Family Secoviridae includes subfamily Comovirinae
- Genus Bacillariornavirus
- Species Kelp fly virus
- Order Tymovirales
- Family Alphaflexiviridae
- Family Betaflexiviridae
- Family Gammaflexiviridae
- Family Tymoviridae
- Unassigned
- Family Alphatetraviridae
- Family Alvernaviridae
- Family Astroviridae
- Family Barnaviridae
- Family Benyviridae
- Family Botourmiaviridae
- Family Bromoviridae
- Family Caliciviridae – includes Norwalk virus
- Family Carmotetraviridae
- Family Closteroviridae
- Family Flaviviridae – includes Yellow fever virus, West Nile virus, Hepatitis C virus, Dengue fever virus, Zika virus
- Family Fusariviridae
- Family Hepeviridae
- Family Hypoviridae
- Family Leviviridae
- Family Luteoviridae – includes Barley yellow dwarf virus
- Family Polycipiviridae
- Family Narnaviridae
- Family Nodaviridae
- Family Permutotetraviridae
- Family Potyviridae
- Family Sarthroviridae
- Family Statovirus
- Family Togaviridae – includes Rubella virus, Ross River virus, Sindbis virus, Chikungunya virus
- Family Tombusviridae
- Family Virgaviridae[34]
- Unassigned genera
- Genus Blunervirus
- Genus Cilevirus
- Genus Higrevirus
- Genus Idaeovirus
- Genus Negevirus
- Genus Ourmiavirus
- Genus Polemovirus
- Genus Sinaivirus
- Genus Sobemovirus
- Unassigned species
- Acyrthosiphon pisum virus
- Bastrovirus
- Blackford virus
- Blueberry necrotic ring blotch virus
- Cadicistrovirus
- Chara australis virus
- Extra small virus
- Goji berry chlorosis virus
- Harmonia axyridis virus 1
- Hepelivirus
- Jingmen tick virus
- Le Blanc virus
- Nedicistrovirus
- Nesidiocoris tenuis virus 1
- Niflavirus
- Nylanderia fulva virus 1
- Orsay virus
- Osedax japonicus RNA virus 1
- Picalivirus
- Planarian secretory cell nidovirus
- Plasmopara halstedii virus
- Rosellinia necatrix fusarivirus 1
- Santeuil virus
- Secalivirus
- Solenopsis invicta virus 3
- Wuhan large pig roundworm virus
Satellite viruses
- Family Sarthroviridae
- Genus Albetovirus
- Genus Aumaivirus
- Genus Papanivirus
- Genus Virtovirus
- Chronic bee paralysis virus
An unclassified astrovirus/hepevirus-like virus has also been described.[35]
Group V – negative-sense ssRNA viruses
[edit]With the exception of the Hepatitis D virus, this group of viruses has been placed into a single phylum—Negarnaviricota. This phylum has been divided into two subphyla—Haploviricotina and Polyploviricotina. Within the subphylum Haploviricotina four classes are currently recognised: Chunqiuviricetes, Milneviricetes, Monjiviricetes and Yunchangviricetes. In the subphylum Polyploviricotina two classes are recognised: Ellioviricetes and Insthoviricetes.
Six classes, seven orders and twenty four families are currently recognized in this group. A number of unassigned species and genera are yet to be classified.[9]
- Phylum Negarnaviricota[36]
- Subphylum Haploviricotina
- Class Chunqiuviricetes
- Order Muvirales
- Family Qinviridae
- Order Muvirales
- Class Milneviricetes
- Order Serpentovirales
- Family Aspiviridae
- Order Serpentovirales
- Class Monjiviricetes
- Order Jingchuvirales
- Family Chuviridae
- Order Mononegavirales
- Family Bornaviridae – Borna disease virus
- Family Filoviridae – includes Ebola virus, Marburg virus
- Family Mymonaviridae
- Family Nyamiviridae[37]
- Family Paramyxoviridae – includes Measles virus, Mumps virus, Nipah virus, Hendra virus, and NDV
- Family Pneumoviridae – includes RSV and Metapneumovirus
- Family Rhabdoviridae – includes Rabies virus
- Family Sunviridae
- Genus Anphevirus
- Genus Arlivirus
- Genus Chengtivirus
- Genus Crustavirus
- Genus Wastrivirus
- Order Jingchuvirales
- Class Yunchangviricetes
- Order Goujianvirales
- Family Yueviridae
- Order Goujianvirales
- Class Chunqiuviricetes
- Subphylum Polyploviricotina
- Class Ellioviricetes
- Order Bunyavirales
- Family Arenaviridae – includes Lassa virus
- Family Cruliviridae
- Family Feraviridae
- Family Fimoviridae
- Family Hantaviridae
- Family Jonviridae
- Family Nairoviridae
- Family Peribunyaviridae
- Family Phasmaviridae
- Family Phenuiviridae
- Family Tospoviridae
- Genus Tilapineviridae
- Order Bunyavirales
- Class Insthoviricetes
- Order Articulavirales
- Family Amnoonviridae – includes Taastrup virus
- Family Orthomyxoviridae – includes Influenza viruses
- Order Articulavirales
- Class Ellioviricetes
- Subphylum Haploviricotina
- Unassigned genera:
- Genus Deltavirus – includes Hepatitis D virus (not a true virus, but a subviral agent)
Gallery
[edit]See also
[edit]- Virus classification
- List of viruses
- Viral replication
- Positive/negative-sense
- Animal viruses
- Double-stranded RNA viruses
- Retrovirus
- DNA viruses
- Norovirus cis-acting replication element
- Viroid
Notes
[edit]- ^ This inclusion was due to TaxoProp 2017.006G, which proposed Riboviria. The confusion might be due to the TaxoProp's reference to a "monophyly of all RNA viruses", improperly termed as it was only demonstrated with RdRP. On the other hand, the proposed definition of Riboviria did correctly mention RdRP .
- ^ The majority of fungal viruses are double-stranded RNA viruses. A small number of positive-strand RNA viruses have been described. One report has suggested the possibility of a negative stranded virus.[7]
References
[edit]- ^ Wagner, Edward K.; Hewlett, Martinez J. (1999). Basic virology. Malden, MA: Blackwell Science, Inc. p. 249. ISBN 0-632-04299-0. Retrieved 30 March 2020.
- ^ International Committee on Taxonomy of Viruses Executive Committee (May 2020). "The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks". Nature Microbiology. 5 (5): 668–674. doi:10.1038/s41564-020-0709-x. PMC 7186216. PMID 32341570.
- ^ TaxoProp 2019.006G
- ^ TaxoProp 2019.009G
- ^ Nguyen M, Haenni AL (June 2003). "Expression strategies of ambisense viruses". Virus Research. 93 (2): 141–50. doi:10.1016/S0168-1702(03)00094-7. PMID 12782362.
- ^ Kondo H, Chiba S, Toyoda K, Suzuki N (January 2013). "Evidence for negative-strand RNA virus infection in fungi". Virology. 435 (2): 201–09. doi:10.1016/j.virol.2012.10.002. PMID 23099204.
- ^ Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R (October 2010). "Viral mutation rates". Journal of Virology. 84 (19): 9733–48. doi:10.1128/JVI.00694-10. PMC 2937809. PMID 20660197.
- ^ a b c Klein DW, Prescott LM, Harley J (1993). Microbiology. Dubuque, Iowa: Wm. C. Brown. ISBN 978-0-697-01372-9.
- ^ Steinhauer DA, Holland JJ (1987). "Rapid evolution of RNA viruses". Annual Review of Microbiology. 41: 409–33. doi:10.1146/annurev.mi.41.100187.002205. PMID 3318675.
- ^ Boutwell CL, Rolland MM, Herbeck JT, Mullins JI, Allen TM (October 2010). "Viral evolution and escape during acute HIV-1 infection". The Journal of Infectious Diseases. 202 (Suppl 2): S309–14. doi:10.1086/655653. PMC 2945609. PMID 20846038.
- ^ Bukh J, Purcell RH, Miller RH (August 1994). "Sequence analysis of the core gene of 14 hepatitis C virus genotypes". Proceedings of the National Academy of Sciences of the United States of America. 91 (17): 8239–43. Bibcode:1994PNAS...91.8239B. doi:10.1073/pnas.91.17.8239. PMC 44581. PMID 8058787.
- ^ Tuplin A, Evans DJ, Simmonds P (October 2004). "Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods". The Journal of General Virology. 85 (Pt 10): 3037–47. doi:10.1099/vir.0.80141-0. PMID 15448367.
- ^ a b Silva JM, Pratas D, Caetano T, Matos D (August 2022). "The complexity landscape of viral genomes". GigaScience. 11: 1–16. doi:10.1093/gigascience/giac079. PMC 9366995. PMID 35950839.
- ^ a b Barr JN, Fearns R (June 2010). "How RNA viruses maintain their genome integrity". The Journal of General Virology. 91 (Pt 6): 1373–87. doi:10.1099/vir.0.020818-0. PMID 20335491.
- ^ Stedman, Kenneth M. (2015). "Deep Recombination: RNA and ssDNA Virus Genes in DNA Virus and Host Genomes". Annual Review of Virology. 2 (1): 203–217. doi:10.1146/annurev-virology-100114-055127. ISSN 2327-0578. PMID 26958913. S2CID 207745438.
- ^ Muslin C, Mac Kain A, Bessaud M, Blondel B, Delpeyroux F (September 2019). "Recombination in Enteroviruses, a Multi-Step Modular Evolutionary Process". Viruses. 11 (9): 859. doi:10.3390/v11090859. PMC 6784155. PMID 31540135.
- ^ Hu WS, Temin HM (November 1990). "Retroviral recombination and reverse transcription". Science. 250 (4985): 1227–33. Bibcode:1990Sci...250.1227H. doi:10.1126/science.1700865. PMID 1700865.
- ^ Rawson JM, Nikolaitchik OA, Keele BF, Pathak VK, Hu WS (November 2018). "Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity". Nucleic Acids Research. 46 (20): 10535–45. doi:10.1093/nar/gky910. PMC 6237782. PMID 30307534.
- ^ a b Bernstein H, Bernstein C, Michod RE (January 2018). "Sex in microbial pathogens". Infection, Genetics and Evolution. 57: 8–25. Bibcode:2018InfGE..57....8B. doi:10.1016/j.meegid.2017.10.024. PMID 29111273.
- ^ a b Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, et al. (June 2016). "Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses". Trends in Microbiology. 24 (6): 490–502. doi:10.1016/j.tim.2016.03.003. PMC 7125511. PMID 27012512.
- ^ Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV (November 2018). "Origins and Evolution of the Global RNA Virome". mBio. 9 (6). doi:10.1128/mBio.02329-18. PMC 6282212. PMID 30482837.
- ^ Francki RI, Fauquet CM, Knudson DL, Brown F (1991). Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses, Archives of Virology (Suppl. 2). Springer. ISBN 978-3-7091-9163-7.
- ^ "Current ICTV Taxonomy Release | ICTV". ictv.global. Retrieved 3 April 2023.
- ^ Koonin EV (September 1991). "The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses". The Journal of General Virology. 72 (Pt 9): 2197–206. doi:10.1099/0022-1317-72-9-2197. PMID 1895057.
- ^ Rozanov MN, Koonin EV, Gorbalenya AE (August 1992). "Conservation of the putative methyltransferase domain: a hallmark of the 'Sindbis-like' supergroup of positive-strand RNA viruses". The Journal of General Virology. 73 (Pt 8): 2129–34. CiteSeerX 10.1.1.532.7367. doi:10.1099/0022-1317-73-8-2129. PMID 1645151.
- ^ Koonin EV, Dolja VV (1993). "Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences". Critical Reviews in Biochemistry and Molecular Biology. 28 (5): 375–430. doi:10.3109/10409239309078440. PMID 8269709.
- ^ Ward CW (1993). "Progress towards a higher taxonomy of viruses". Research in Virology. 144 (6): 419–53. doi:10.1016/S0923-2516(06)80059-2. PMC 7135741. PMID 8140287.
- ^ Koonin EV, Wolf YI, Nagasaki K, Dolja VV (December 2008). "The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups". Nature Reviews. Microbiology. 6 (12): 925–39. doi:10.1038/nrmicro2030. PMID 18997823.
- ^ a b Ghabrial SA (1998). "Origin, adaptation and evolutionary pathways of fungal viruses". Virus Genes. 16 (1): 119–31. doi:10.1023/a:1007966229595. PMC 7089520. PMID 9562896.
- ^ Gibrat JF, Mariadassou M, Boudinot P, Delmas B (July 2013). "Analyses of the radiation of birnaviruses from diverse host phyla and of their evolutionary affinities with other double-stranded RNA and positive strand RNA viruses using robust structure-based multiple sequence alignments and advanced phylogenetic methods". BMC Evolutionary Biology. 13 (1): 154. Bibcode:2013BMCEE..13..154G. doi:10.1186/1471-2148-13-154. PMC 3724706. PMID 23865988.
- ^ Li CX, Shi M, Tian JH, Lin XD, Kang YJ, Chen LJ, et al. (January 2015). "Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses". eLife. 4. doi:10.7554/eLife.05378. PMC 4384744. PMID 25633976.
- ^ Krupovic M, Kuhn JH, Fischer MG (January 2016). "A classification system for virophages and satellite viruses". Archives of Virology. 161 (1): 233–47. doi:10.1007/s00705-015-2622-9. hdl:11858/00-001M-0000-0028-DC34-F. PMID 26446887.
- ^ Adams MJ, Antoniw JF, Kreuze J (2009). "Virgaviridae: a new family of rod-shaped plant viruses". Archives of Virology. 154 (12): 1967–72. doi:10.1007/s00705-009-0506-6. PMID 19862474.
- ^ Pankovics P, Boros Á, Kiss T, Engelmann P, Reuter G (2019) Genetically highly divergent RNA virus with astrovirus-like (5'-end) and hepevirus-like (3'-end) genome organization in carnivorous birds, European roller (Coracias garrulus). Infect Genet Evol
- ^ "Virus Taxonomy: 2018 Release". International Committee on Taxonomy of Viruses. Retrieved 13 November 2018.
- ^ Mihindukulasuriya KA, Nguyen NL, Wu G, Huang HV, da Rosa AP, Popov VL, et al. (May 2009). "Nyamanini and midway viruses define a novel taxon of RNA viruses in the order Mononegavirales". Journal of Virology. 83 (10): 5109–16. doi:10.1128/JVI.02667-08. PMC 2682064. PMID 19279111.
External links
[edit]- RNA Viruses at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Animal viruses