Human mitochondrial genetics
| Human mitochondrial DNA | |
|---|---|
 The 16,569 bp long human mitochondrial genome with the protein-coding (red, orange, yellow), ribosomal RNA (blue), and transfer RNA genes (white). Non-coding mtDNA control region in grey. | |
| Features | |
| Length (bp) | 16,569 |
| No. of genes | 13 (coding genes) 24 (non coding genes) |
| Type | Mitochondrial DNA |
| Complete gene lists | |
| HGNC | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome MT |
| Entrez | Chromosome MT |
| NCBI | Chromosome MT |
| UCSC | Chromosome M |
| Full DNA sequences | |
| RefSeq | NC_012920 (FASTA) |
| GenBank | J01415 (FASTA) |

Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA (the DNA contained in human mitochondria). The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondria are small structures in cells that generate energy for the cell to use, and are hence referred to as the "powerhouses" of the cell.
Mitochondrial DNA (mtDNA) is not transmitted through nuclear DNA (nDNA). In humans, as in most multicellular organisms, mitochondrial DNA is inherited only from the mother's ovum. There are theories, however, that paternal mtDNA transmission in humans can occur under certain circumstances.[3] Mitochondrial inheritance is therefore non-Mendelian, as Mendelian inheritance presumes that half the genetic material of a fertilized egg (zygote) derives from each parent.
This allowed the creation of mitochondrial DNA haplogroups to study population genetics.
Eighty percent of mitochondrial DNA codes for mitochondrial RNA, and therefore most mitochondrial DNA mutations lead to functional problems, which may be manifested as muscle disorders (myopathies).
Because they provide 30 molecules of ATP per glucose molecule in contrast to the 2 ATP molecules produced by glycolysis, mitochondria are essential to all higher organisms for sustaining life. The mitochondrial diseases are genetic disorders carried in mitochondrial DNA, or nuclear DNA coding for mitochondrial components. Slight problems with any one of the numerous enzymes used by the mitochondria can be devastating to the cell, and in turn, to the organism.
Quantity
[edit]In humans, mitochondrial DNA (mtDNA) forms closed circular molecules that contain 16,569[4][5] DNA base pairs,[6] with each such molecule normally containing a full set of the mitochondrial genes. Each human mitochondrion contains, on average, approximately 5 such mtDNA molecules, with the quantity ranging between 1 and 15.[6] Each human cell contains approximately 100 mitochondria, giving a total number of mtDNA molecules per human cell of approximately 500.[6] The amount of mitochondria per cell also varies by cell type, with some examples being:
- Erythrocytes: 0 mitochondria per cell.[1]
- Lymphocytes: 3 mitochondria per cell.[7]
- Egg cell: Mature metaphase II egg cells can contain 100,000 mitochondria, and 50,000–1,500,000 copies of the mitochondrial genome (corresponding to up to 90% of the egg cell DNA).[2]
Inheritance patterns
[edit]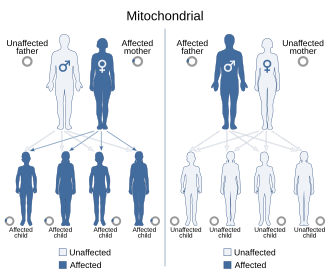

Because mitochondrial diseases (diseases due to malfunction of mitochondria) can be inherited both maternally and through chromosomal inheritance, the way in which they are passed on from generation to generation can vary greatly depending on the disease. Mitochondrial genetic mutations that occur in the nuclear DNA can occur in any of the chromosomes (depending on the species). Mutations inherited through the chromosomes can be autosomal dominant or recessive and can also be sex-linked dominant or recessive. Chromosomal inheritance follows normal Mendelian laws, despite the fact that the phenotype of the disease may be masked.
Because of the complex ways in which mitochondrial and nuclear DNA "communicate" and interact, even seemingly simple inheritance is hard to diagnose. A mutation in chromosomal DNA may change a protein that regulates (increases or decreases) the production of another certain protein in the mitochondria or the cytoplasm; this may lead to slight, if any, noticeable symptoms. On the other hand, some devastating mtDNA mutations are easy to diagnose because of their widespread damage to muscular, neural, and/or hepatic tissues (among other high-energy and metabolism-dependent tissues) and because they are present in the mother and all the offspring.
The number of affected mtDNA molecules inherited by a specific offspring can vary greatly because
- the mitochondria within the fertilized oocyte is what the new life will have to begin with (in terms of mtDNA),
- the number of affected mitochondria varies from cell (in this case, the fertilized oocyte) to cell depending both on the number it inherited from its mother cell and environmental factors which may favor mutant or wildtype mitochondrial DNA,
- the number of mtDNA molecules in the mitochondria varies from around two to ten.
It is possible, even in twin births, for one baby to receive more than half mutant mtDNA molecules while the other twin may receive only a tiny fraction of mutant mtDNA molecules with respect to wildtype (depending on how the twins divide from each other and how many mutant mitochondria happen to be on each side of the division). In a few cases, some mitochondria or a mitochondrion from the sperm cell enters the oocyte but paternal mitochondria are actively decomposed.
Genes
[edit]Genes in the human mitochondrial genome are as follows.
Electron transport chain, and humanin
[edit]It was originally incorrectly believed that the mitochondrial genome contained only 13 protein-coding genes, all of them encoding proteins of the electron transport chain. However, in 2001, a 14th biologically active protein called humanin was discovered, and was found to be encoded by the mitochondrial gene MT-RNR2 which also encodes part of the mitochondrial ribosome (made out of RNA):
| Complex number |
Category | Genes | Positions in the mitogenome | Strand |
|---|---|---|---|---|
| I | NADH dehydrogenase | |||
| MT-ND1 | 3,307–4,262 | H | ||
| MT-ND2 | 4,470–5,511 | H | ||
| MT-ND3 | 10,059–10,404 | H | ||
| MT-ND4L | 10,470–10,766 | H | ||
| MT-ND4 | 10,760–12,137 (overlap with MT-ND4L) | H | ||
| MT-ND5 | 12,337–14,148 | H | ||
| MT-ND6 | 14,149–14,673 | L | ||
| III | Coenzyme Q - cytochrome c reductase / Cytochrome b | MT-CYB | 14,747–15,887 | H |
| IV | Cytochrome c oxidase | MT-CO1 | 5,904–7,445 | H |
| MT-CO2 | 7,586–8,269 | H | ||
| MT-CO3 | 9,207–9,990 | H | ||
| V | ATP synthase | MT-ATP6 | 8,527–9,207 (overlap with MT-ATP8) | H |
| MT-ATP8 | 8,366–8,572 | H | ||
| — | Humanin | MT-RNR2 | — | — |
Unlike the other proteins, humanin does not remain in the mitochondria, and interacts with the rest of the cell and cellular receptors. Humanin can protect brain cells by inhibiting apoptosis. Despite its name, versions of humanin also exist in other animals, such as rattin in rats.
rRNA
[edit]The following genes encode rRNAs:
| Subunit | rRNA | Genes | Positions in the mitogenome | Strand |
|---|---|---|---|---|
| Small (SSU) | 12S | MT-RNR1 | 648–1,601 | H |
| Large (LSU) | 16S | MT-RNR2 | 1,671–3,229 | H |
tRNA
[edit]The following genes encode tRNAs:
| Amino Acid | 3-Letter | 1-Letter | MT DNA | Positions | Strand |
|---|---|---|---|---|---|
| Alanine | Ala | A | MT-TA | 5,587–5,655 | L |
| Arginine | Arg | R | MT-TR | 10,405–10,469 | H |
| Asparagine | Asn | N | MT-TN | 5,657–5,729 | L |
| Aspartic acid | Asp | D | MT-TD | 7,518–7,585 | H |
| Cysteine | Cys | C | MT-TC | 5,761–5,826 | L |
| Glutamic acid | Glu | E | MT-TE | 14,674–14,742 | L |
| Glutamine | Gln | Q | MT-TQ | 4,329–4,400 | L |
| Glycine | Gly | G | MT-TG | 9,991–10,058 | H |
| Histidine | His | H | MT-TH | 12,138–12,206 | H |
| Isoleucine | Ile | I | MT-TI | 4,263–4,331 | H |
| Leucine | Leu (UUR) | L | MT-TL1 | 3,230–3,304 | H |
| Leucine | Leu (CUN) | L | MT-TL2 | 12,266–12,336 | H |
| Lysine | Lys | K | MT-TK | 8,295–8,364 | H |
| Methionine | Met | M | MT-TM | 4,402–4,469 | H |
| Phenylalanine | Phe | F | MT-TF | 577–647 | H |
| Proline | Pro | P | MT-TP | 15,956–16,023 | L |
| Serine | Ser (UCN) | S | MT-TS1 | 7,446–7,514 | L |
| Serine | Ser (AGY) | S | MT-TS2 | 12,207–12,265 | H |
| Threonine | Thr | T | MT-TT | 15,888–15,953 | H |
| Tryptophan | Trp | W | MT-TW | 5,512–5,579 | H |
| Tyrosine | Tyr | Y | MT-TY | 5,826–5,891 | L |
| Valine | Val | V | MT-TV | 1,602–1,670 | H |
Location of genes
[edit]Mitochondrial DNA traditionally had the two strands of DNA designated the heavy and the light strand, due to their buoyant densities during separation in cesium chloride gradients,[8][9] which was found to be related to the relative G+T nucleotide content of the strand.[10] However, confusion of labeling of this strands is widespread, and appears to originate with an identification of the majority coding strand as the heavy in one influential article in 1999.[11][10] In humans, the light strand of mtDNA carries 28 genes and the heavy strand of mtDNA carries only 9 genes.[10][12] Eight of the 9 genes on the heavy strand code for mitochondrial tRNA molecules. Human mtDNA consists of 16,569 nucleotide pairs. The entire molecule is regulated by only one regulatory region which contains the origins of replication of both heavy and light strands. The entire human mitochondrial DNA molecule has been mapped[1][2].
Genetic code variants
[edit]The genetic code is, for the most part, universal, with few exceptions:[13] mitochondrial genetics includes some of these. For most organisms the "stop codons" are "UAA", "UAG", and "UGA". In vertebrate mitochondria "AGA" and "AGG" are also stop codons, but not "UGA", which codes for tryptophan instead. "AUA" codes for isoleucine in most organisms but for methionine in vertebrate mitochondrial mRNA.
There are many other variations among the codes used by other mitochondrial m/tRNA, which happened not to be harmful to their organisms, and which can be used as a tool (along with other mutations among the mtDNA/RNA of different species) to determine relative proximity of common ancestry of related species. (The more related two species are, the more mtDNA/RNA mutations will be the same in their mitochondrial genome).
Using these techniques, it is estimated that the first mitochondria arose around 1.5 billion years ago. A generally accepted hypothesis is that mitochondria originated as an aerobic prokaryote in a symbiotic relationship within an anaerobic eukaryote.
Replication, repair, transcription, and translation
[edit]Mitochondrial replication is controlled by nuclear genes and is specifically suited to make as many mitochondria as that particular cell needs at the time.
Mitochondrial transcription in humans is initiated from three promoters, H1, H2, and L (heavy strand 1, heavy strand 2, and light strand promoters). The H2 promoter transcribes almost the entire heavy strand and the L promoter transcribes the entire light strand. The H1 promoter causes the transcription of the two mitochondrial rRNA molecules.[14]
When transcription takes place on the heavy strand a polycistronic transcript is created. The light strand produces either small transcripts, which can be used as primers, or one long transcript. The production of primers occurs by processing of light strand transcripts with the Mitochondrial RNase MRP (Mitochondrial RNA Processing). The requirement of transcription to produce primers links the process of transcription to mtDNA replication. Full length transcripts are cut into functional tRNA, rRNA, and mRNA molecules.[citation needed]
The process of transcription initiation in mitochondria involves three types of proteins: the mitochondrial RNA polymerase (POLRMT), mitochondrial transcription factor A (TFAM), and mitochondrial transcription factors B1 and B2 (TFB1M, TFB2M). POLRMT, TFAM, and TFB1M or TFB2M assemble at the mitochondrial promoters and begin transcription. The actual molecular events that are involved in initiation are unknown, but these factors make up the basal transcription machinery and have been shown to function in vitro.[citation needed]
Mitochondrial translation is still not very well understood. In vitro translations have still not been successful, probably due to the difficulty of isolating sufficient mt mRNA, functional mt rRNA, and possibly because of the complicated changes that the mRNA undergoes before it is translated.[citation needed]
Mitochondrial DNA polymerase
[edit]The Mitochondrial DNA Polymerase (Pol gamma, encoded by the POLG gene) is used in the copying of mtDNA during replication. Because the two (heavy and light) strands on the circular mtDNA molecule have different origins of replication, it replicates in a D-loop mode. One strand begins to replicate first, displacing the other strand. This continues until replication reaches the origin of replication on the other strand, at which point the other strand begins replicating in the opposite direction. This results in two new mtDNA molecules. Each mitochondrion has several copies of the mtDNA molecule and the number of mtDNA molecules is a limiting factor in mitochondrial fission. After the mitochondrion has enough mtDNA, membrane area, and membrane proteins, it can undergo fission (very similar to that which bacteria use) to become two mitochondria. Evidence suggests that mitochondria can also undergo fusion and exchange (in a form of crossover) genetic material among each other. Mitochondria sometimes form large matrices in which fusion, fission, and protein exchanges are constantly occurring. mtDNA shared among mitochondria (despite the fact that they can undergo fusion).[citation needed]
Damage and transcription error
[edit]Mitochondrial DNA is susceptible to damage from free oxygen radicals from mistakes that occur during the production of ATP through the electron transport chain. These mistakes can be caused by genetic disorders, cancer, and temperature variations. These radicals can damage mtDNA molecules or change them, making it hard for mitochondrial polymerase to replicate them. Both cases can lead to deletions, rearrangements, and other mutations. Recent evidence has suggested that mitochondria have enzymes that proofread mtDNA and fix mutations that may occur due to free radicals. It is believed that a DNA recombinase found in mammalian cells is also involved in a repairing recombination process. Deletions and mutations due to free radicals have been associated with the aging process. It is believed that radicals cause mutations which lead to mutant proteins, which in turn led to more radicals. This process takes many years and is associated with some aging processes involved in oxygen-dependent tissues such as brain, heart, muscle, and kidney. Auto-enhancing processes such as these are possible causes of degenerative diseases including Parkinson's, Alzheimer's, and coronary artery disease.[citation needed]
Chromosomally mediated mtDNA replication errors
[edit]Because mitochondrial growth and fission are mediated by the nuclear DNA, mutations in nuclear DNA can have a wide array of effects on mtDNA replication. Despite the fact that the loci for some of these mutations have been found on human chromosomes, specific genes and proteins involved have not yet been isolated. Mitochondria need a certain protein to undergo fission. If this protein (generated by the nucleus) is not present, the mitochondria grow but they do not divide. This leads to giant, inefficient mitochondria. Mistakes in chromosomal genes or their products can also affect mitochondrial replication more directly by inhibiting mitochondrial polymerase and can even cause mutations in the mtDNA directly and indirectly. Indirect mutations are most often caused by radicals created by defective proteins made from nuclear DNA.[citation needed]
Mitochondrial diseases
[edit]Contribution of mitochondrial versus nuclear genome
[edit]In total, the mitochondrion hosts about 3000 different types of proteins, but only about 13 of them are coded on the mitochondrial DNA. Most of the 3000 types of proteins are involved in a variety of processes other than ATP production, such as porphyrin synthesis. Only about 3% of them code for ATP production proteins. This means most of the genetic information coding for the protein makeup of mitochondria is in chromosomal DNA and is involved in processes other than ATP synthesis. This increases the chances that a mutation that will affect a mitochondrion will occur in chromosomal DNA, which is inherited in a Mendelian pattern. Another result is that a chromosomal mutation will affect a specific tissue due to its specific needs, whether those may be high energy requirements or a need for the catabolism or anabolism of a specific neurotransmitter or nucleic acid. Because several copies of the mitochondrial genome are carried by each mitochondrion (2–10 in humans), mitochondrial mutations can be inherited maternally by mtDNA mutations which are present in mitochondria inside the oocyte before fertilization, or (as stated above) through mutations in the chromosomes.[citation needed]
Presentation
[edit]Mitochondrial diseases range in severity from asymptomatic to fatal, and are most commonly due to inherited rather than acquired mutations of mitochondrial DNA. A given mitochondrial mutation can cause various diseases depending on the severity of the problem in the mitochondria and the tissue the affected mitochondria are in. Conversely, several different mutations may present themselves as the same disease. This almost patient-specific characterization of mitochondrial diseases (see Personalized medicine) makes them very hard to accurately recognize, diagnose and trace. Some diseases are observable at or even before birth (many causing death) while others do not show themselves until late adulthood (late-onset disorders). This is because the number of mutant versus wildtype mitochondria varies between cells and tissues, and is continuously changing. Because cells have multiple mitochondria, different mitochondria in the same cell can have different variations of the mtDNA. This condition is referred to as heteroplasmy. When a certain tissue reaches a certain ratio of mutant versus wildtype mitochondria, a disease will present itself. The ratio varies from person to person and tissue to tissue (depending on its specific energy, oxygen, and metabolism requirements, and the effects of the specific mutation). Mitochondrial diseases are very numerous and different. Apart from diseases caused by abnormalities in mitochondrial DNA, many diseases are suspected to be associated in part by mitochondrial dysfunctions, such as diabetes mellitus,[15] forms of cancer[16] and cardiovascular disease, lactic acidosis,[17] specific forms of myopathy,[18] osteoporosis,[19] Alzheimer's disease,[20] Parkinsons's disease,[21] stroke,[22] male infertility[23] and which are also believed to play a role in the aging process.[24]
Use in forensics
[edit]Human mtDNA can also be used to help identify individuals.[25] Forensic laboratories occasionally use mtDNA comparison to identify human remains, and especially to identify older unidentified skeletal remains. Although unlike nuclear DNA, mtDNA is not specific to one individual, it can be used in combination with other evidence (anthropological evidence, circumstantial evidence, and the like) to establish identification. mtDNA is also used to exclude possible matches between missing persons and unidentified remains.[26] Many researchers believe that mtDNA is better suited to identification of older skeletal remains than nuclear DNA because the greater number of copies of mtDNA per cell increases the chance of obtaining a useful sample, and because a match with a living relative is possible even if numerous maternal generations separate the two.
Examples
[edit]American outlaw Jesse James's remains were identified using a comparison between mtDNA extracted from his remains and the mtDNA of the son of the female-line great-granddaughter of his sister.[27]
Similarly, the remains of Alexandra Feodorovna (Alix of Hesse), last Empress of Russia, and her children were identified by comparison of their mitochondrial DNA with that of Prince Philip, Duke of Edinburgh, whose maternal grandmother was Alexandra's sister Victoria of Hesse.[28]
Similarly to identify Emperor Nicholas II remains his mitochondrial DNA was compared with that of James Carnegie, 3rd Duke of Fife, whose maternal great-grandmother Alexandra of Denmark (Queen Alexandra) was sister of Nicholas II mother Dagmar of Denmark (Empress Maria Feodorovna).[28][29]
Similarly were the remains of King Richard III identified.[30]
See also
[edit]- Paternal mtDNA transmission
- Human mitochondrial DNA haplogroups
- Cambridge Reference Sequence
- Human mitochondrial molecular clock
- Genetic genealogy for lists of databases which help users find others with their Y-DNA and mtDNA.
References
[edit]- ^ a b Shuster RC, Rubenstein AJ, Wallace DC (1988). "Mitochondrial DNA in anucleate human blood cells". Biochem Biophys Res Commun. 155 (3): 1360–5. doi:10.1016/s0006-291x(88)81291-9. PMID 3178814.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Zhang D, Keilty D, Zhang ZF, Chian RC (2017). "Mitochondria in oocyte aging: current understanding". Facts Views Vis Obgyn. 9 (1): 29–38. PMC 5506767. PMID 28721182.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Schwartz, Marianne; Vissing, John (22 August 2002). "Paternal Inheritance of Mitochondrial DNA". New England Journal of Medicine. 347 (8): 576–580. doi:10.1056/NEJMoa020350. PMID 12192017.
- ^ Anderson, S.; Bankier, A. T.; Barrell, B. G.; de Bruijn, M. H. L.; Coulson, A. R.; Drouin, J.; Eperon, I. C.; Nierlich, D. P.; Roe, B. A.; Sanger, F.; Schreier, P. H.; Smith, A. J. H.; Staden, R.; Young, I. G. (April 1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–465. Bibcode:1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534. S2CID 4355527.
- ^ "Untitled". Archived from the original on 2011-08-13. Retrieved 2012-06-13.
- ^ a b c Satoh, M; Kuroiwa, T (September 1991). "Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell". Experimental Cell Research. 196 (1): 137–140. doi:10.1016/0014-4827(91)90467-9. PMID 1715276.
- ^ Nagy G, Barcza M, Gonchoroff N, Phillips PE, Perl A (2004). "Nitric oxide-dependent mitochondrial biogenesis generates Ca2+ signaling profile of lupus T cells". J Immunol. 173 (6): 3676–83. doi:10.4049/jimmunol.173.6.3676. PMC 4034140. PMID 15356113.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zimmerman, Earl G.; Akins, Darrin R.; Planz, John V.; Schurr, Michael J. (September 1988). "A rapid procedure for isolating mitochondrial DNA". Gene Analysis Techniques. 5 (5): 102–104. doi:10.1016/0735-0651(88)90004-0. PMID 2847966.
- ^ Welter, Cornelius; Meese, Eckart; Blin, Nikolaus (1988). "Rapid step-gradient purification of mitochondrial DNA". Molecular Biology Reports. 13 (2): 117–120. doi:10.1007/BF00539059. PMID 3221842. S2CID 3157709.
- ^ a b c Barroso Lima, Nicholas Costa; Prosdocimi, Francisco (17 February 2018). "The heavy strand dilemma of vertebrate mitochondria on genome sequencing age: number of encoded genes or G + T content?". Mitochondrial DNA Part A. 29 (2): 300–302. doi:10.1080/24701394.2016.1275603. PMID 28129726. S2CID 20552678.
- ^ Taanman, Jan-Willem (February 1999). "The mitochondrial genome: structure, transcription, translation and replication". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1410 (2): 103–123. doi:10.1016/s0005-2728(98)00161-3. PMID 10076021.
- ^ Anderson, S.; Bankier, A. T.; Barrell, B. G.; de Bruijn, M. H. L.; Coulson, A. R.; Drouin, J.; Eperon, I. C.; Nierlich, D. P.; Roe, B. A.; Sanger, F.; Schreier, P. H.; Smith, A. J. H.; Staden, R.; Young, I. G. (1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–65. Bibcode:1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534. S2CID 4355527.
- ^ "The Genetic Codes". www.ncbi.nlm.nih.gov. National Center for Biotechnology Information. Retrieved 16 March 2019.
- ^ Asin-Cayuela, Jordi; Gustafsson, Claes M. (2007). "Mitochondrial transcription and its regulation in mammalian cells". Trends in Biochemical Sciences. 32 (3): 111–17. doi:10.1016/j.tibs.2007.01.003. PMID 17291767.
- ^ Tanaka, Masashi; Fuku, Noriyuki; Nishigaki, Yutaka; Matsuo, Hitoshi; Segawa, Tomonori; Watanabe, Sachiro; Kato, Kimihiko; Yoko, Kiyoshi; Ito, Masafumi; Nozawa, Yoshinori; Yamada, Yoshiji (February 2007). "Women With Mitochondrial Haplogroup N9a Are Protected Against Metabolic Syndrome". Diabetes. 56 (2): 518–521. doi:10.2337/db06-1105. PMID 17259400. S2CID 34199769.
- ^ Theodoratou, Evropi; Din, Farhat V.N.; Farrington, Susan M.; Cetnarskyj, Roseanne; Barnetson, Rebecca A.; Porteous, Mary E.; Dunlop, Malcolm G.; Campbell, Harry; Tenesa, Albert (February 2010). "Association between common mtDNA variants and all-cause or colorectal cancer mortality". Carcinogenesis. 31 (2): 296–301. doi:10.1093/carcin/bgp237. PMID 19945968.
- ^ Goto, Y (September 1993). "[MELAS (mitochondrial myopathy, encephalopathy lactic acidosis, and stroke-like episodes): clinical features and mitochondrial DNA mutations]". Nihon Rinsho. Japanese Journal of Clinical Medicine. 51 (9): 2373–8. PMID 8411715.
- ^ Ahuja, Abhimanyu S. (21 May 2018). "Understanding mitochondrial myopathies: a review". PeerJ. 6: e4790. doi:10.7717/peerj.4790. PMC 5967365. PMID 29844960.
- ^ Angireddy, Rajesh; Kazmi, Hasan Raza; Srinivasan, Satish; Sun, Li; Iqbal, Jameel; Fuchs, Serge Y.; Guha, Manti; Kijima, Takashi; Yuen, Tony; Zaidi, Mone; Avadhani, Narayan G. (August 2019). "Cytochrome c oxidase dysfunction enhances phagocytic function and osteoclast formation in macrophages". The FASEB Journal. 33 (8): 9167–9181. doi:10.1096/fj.201900010RR. PMC 6662975. PMID 31063702.
- ^ Carrieri, Giuseppina; Bonafè, Massimiliano; De Luca, Maria; Rose, Giuseppina; Varcasia, Ottavia; Bruni, Amalia; Maletta, Raffaele; Nacmias, Benedetta; Sorbi, Sandro; Corsonello, Francesco; Feraco, Emidio; Andreev, Kirill F.; Yashin, Anatoli I.; Franceschi, Claudio; De Benedictis, Giovanna (March 2001). "Mitochondrial DNA haplogroups and APOE4 allele are non-independent variables in sporadic Alzheimer's disease". Human Genetics. 108 (3): 194–198. doi:10.1007/s004390100463. PMID 11354629. S2CID 6171041.
- ^ Martín-Jiménez, Rebeca; Lurette, Olivier; Hebert-Chatelain, Etienne (1 August 2020). "Damage in Mitochondrial DNA Associated with Parkinson's Disease". DNA and Cell Biology. 39 (8): 1421–1430. doi:10.1089/dna.2020.5398. PMID 32397749.
- ^ Chinnery, Patrick F; Elliott, Hannah R; Syed, Anila; Rothwell, Peter M (May 2010). "Mitochondrial DNA haplogroups and risk of transient ischaemic attack and ischaemic stroke: a genetic association study". The Lancet Neurology. 9 (5): 498–503. doi:10.1016/S1474-4422(10)70083-1. PMC 2855429. PMID 20362514.
- ^ Ruiz-Pesini, Eduardo; Lapeña, Ana-Cristina; Díez-Sánchez, Carmen; Pérez-Martos, Acisclo; Montoya, Julio; Alvarez, Enrique; Díaz, Miguel; Urriés, Antonio; Montoro, Luis; López-Pérez, Manuel J.; Enríquez, José A. (September 2000). "Human mtDNA Haplogroups Associated with High or Reduced Spermatozoa Motility". The American Journal of Human Genetics. 67 (3): 682–696. doi:10.1086/303040. PMC 1287528. PMID 10936107.
- ^ Courtenay, Monique D.; Gilbert, John R.; Jiang, Lan; Cummings, Anna C.; Gallins, Paul J.; Caywood, Laura; Reinhart-Mercer, Lori; Fuzzell, Denise; Knebusch, Claire; Laux, Renee; McCauley, Jacob L.; Jackson, Charles E.; Pericak-Vance, Margaret A.; Haines, Jonathan L.; Scott, William K. (February 2012). "Mitochondrial Haplogroup X is associated with successful aging in the Amish". Human Genetics. 131 (2): 201–208. doi:10.1007/s00439-011-1060-3. PMC 4834861. PMID 21750925.
- ^ Brown, W. M. (1 June 1980). "Polymorphism in mitochondrial DNA of humans as revealed by restriction endonuclease analysis". Proceedings of the National Academy of Sciences. 77 (6): 3605–3609. Bibcode:1980PNAS...77.3605B. doi:10.1073/pnas.77.6.3605. PMC 349666. PMID 6251473.
- ^ "Paleo-DNA Laboratory – Forensic Services". Archived from the original on 2012-03-13. Retrieved 2012-06-13.
- ^ Stone, Anne C.; Starrs, James E.; Stoneking, Mark (1 January 2001). "Mitochondrial DNA Analysis of the Presumptive Remains of Jesse James". Journal of Forensic Sciences. 46 (1): 173–6. doi:10.1520/JFS14932J. PMID 11210907. S2CID 6480921.
- ^ a b Gill, Peter; Ivanov, Pavel L.; Kimpton, Colin; Piercy, Romelle; Benson, Nicola; Tully, Gillian; Evett, Ian; Hagelberg, Erika; Sullivan, Kevin (February 1994). "Identification of the remains of the Romanov family by DNA analysis". Nature Genetics. 6 (2): 130–135. doi:10.1038/ng0294-130. PMID 8162066. S2CID 33557869.
- ^ Ivanov, Pavel L.; Wadhams, Mark J.; Roby, Rhonda K.; Holland, Mitchell M.; Weedn, Victor W.; Parsons, Thomas J. (April 1996). "Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II". Nature Genetics. 12 (4): 417–420. doi:10.1038/ng0496-417. PMID 8630496. S2CID 287478.
- ^ Ashdown-Hill, John (2013). The Last Days of Richard III and the Fate of His DNA. History Press. ISBN 978-0-7524-9205-6.[page needed]
Further reading
[edit]- Li, Xiangqi; Liu, Lianyong; Xi, Qian; Zhao, Xuemei; Fang, Mingshuang; Ma, Junhua; Zhu, Zhaohui; Wang, Xing; Shi, Chao; Wang, Jingnan; Zhu, Hongling; Zhang, Jichen; Zhang, Chaobao; Hu, Shuanggang; Ni, Minjie; Gu, Mingjun (2016). "Short-term serum deprivation causes no significant mitochondrial DNA mutation in vascular smooth muscle cells revealed by a new next generation sequencing technology". Acta Biochimica et Biophysica Sinica. 48 (9): 862–4. doi:10.1093/abbs/gmw059. PMID 27261779.
External links
[edit]- ISOGG YBrowse: A genome browser.
- mitoWheel: A mitogenome browser.
- Mitochondrial DNA in MedlinePlus
- MITOMAP, reports on published data about human mitochondrial DNA variation
