Marine viruses

Marine viruses are defined by their habitat as viruses that are found in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Viruses are small infectious agents that can only replicate inside the living cells of a host organism, because they need the replication machinery of the host to do so.[4] They can infect all types of life forms, from animals and plants to microorganisms, including bacteria and archaea.[5]
When not inside a cell or in the process of infecting a cell, viruses exist in the form of independent particles called virions. A virion contains a genome (a long molecule that carries genetic information in the form of either DNA or RNA) surrounded by a capsid (a protein coat protecting the genetic material). The shapes of these virus particles range from simple helical and icosahedral forms for some virus species to more complex structures for others. Most virus species have virions that are too small to be seen with an optical microscope. The average virion is about one one-hundredth the linear size of the average bacterium.
A teaspoon of seawater typically contains about fifty million viruses.[6] Most of these viruses are bacteriophages which infect and destroy marine bacteria and control the growth of phytoplankton at the base of the marine food web. Bacteriophages are harmless to plants and animals but are essential to the regulation of marine ecosystems. They supply key mechanisms for recycling ocean carbon and nutrients. In a process known as the viral shunt, organic molecules released from dead bacterial cells stimulate fresh bacterial and algal growth. In particular, the breaking down of bacteria by viruses (lysis) has been shown to enhance nitrogen cycling and stimulate phytoplankton growth. Viral activity also affects the biological pump, the process which sequesters carbon in the deep ocean. By increasing the amount of respiration in the oceans, viruses are indirectly responsible for reducing the amount of carbon dioxide in the atmosphere by approximately 3 gigatonnes of carbon per year.
Marine microorganisms make up about 70% of the total marine biomass. It is estimated marine viruses kill 20% of the microorganism biomass every day. Viruses are the main agents responsible for the rapid destruction of harmful algal blooms which often kill other marine life. The number of viruses in the oceans decreases further offshore and deeper into the water, where there are fewer host organisms. Viruses are an important natural means of transferring genes between different species, which increases genetic diversity and drives evolution. It is thought viruses played a central role in early evolution before the diversification of bacteria, archaea and eukaryotes, at the time of the last universal common ancestor of life on Earth. Viruses are still one of the largest areas of unexplored genetic diversity on Earth.
| Part of a series of overviews on |
| Marine life |
|---|
 |
Background
[edit]| Part of a series on |
| Plankton |
|---|
 |
Viruses are now recognised as ancient and as having origins that pre-date the divergence of life into the three domains.[7] They are found wherever there is life and have probably existed since living cells first evolved.[8] The origins of viruses in the evolutionary history of life are unclear because they do not form fossils. Molecular techniques are used to compare the DNA or RNA of viruses and are a useful means of investigating how they arose.[9] Some viruses may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity.[10]
Opinions differ on whether viruses are a form of life or organic structures that interact with living organisms.[11] They are considered by some to be a life form, because they carry genetic material, reproduce by creating multiple copies of themselves through self-assembly, and evolve through natural selection. However they lack key characteristics such as a cellular structure generally considered necessary to count as life. Because they possess some but not all such qualities, viruses have been described as replicators[12] and as "organisms at the edge of life".[13]
The existence of viruses in the ocean was discovered through electron microscopy and epifluorescence microscopy of ecological water samples, and later through metagenomic sampling of uncultured viral samples.[14][15] Marine viruses, although microscopic and essentially unnoticed by scientists until recently, are the most abundant and diverse biological entities in the ocean. Viruses have an estimated abundance of 1030 in the ocean, or between 106 and 1011 viruses per millilitre.[4] Quantification of marine viruses was originally performed using transmission electron microscopy but has been replaced by epifluorescence or flow cytometry.[16]
Bacteriophages
[edit]
Bacteriophages, often contracted to phages, are viruses that parasitize bacteria for replication. As aptly named, marine phages parasitize marine bacteria, such as cyanobacteria.[17] They are a diverse group of viruses which are the most abundant biological entity in marine environments, because their hosts, bacteria, are typically the numerically dominant cellular life in the sea. There are up to ten times more phages in the oceans than there are bacteria,[18] reaching levels of 250 million bacteriophages per millilitre of seawater.[19] These viruses infect specific bacteria by binding to surface receptor molecules and then entering the cell. Within a short amount of time, in some cases just minutes, bacterial polymerase starts translating viral mRNA into protein. These proteins go on to become either new virions within the cell, helper proteins, which help assembly of new virions, or proteins involved in cell lysis. Viral enzymes aid in the breakdown of the cell membrane, and there are phages that can replicate three hundred phages twenty minutes after injection.[20]

interacting with the marine Prochlorococcus MED4 bacterium

(b) same image visualised by highlighting the cell wall in orange, the plasma membrane in light yellow, the thylakoid membrane in green, carboxysomes in cyan, the polyphosphate body in blue, adsorbed phages on the sides or top of the cell in red, and cytoplasmic granules (probably mostly ribosomes) in light purple.[21]
Bacteria defend themselves from bacteriophages by producing enzymes that destroy foreign DNA. These enzymes, called restriction endonucleases, cut up the viral DNA that bacteriophages inject into bacterial cells.[22] Bacteria also contain a system that uses CRISPR sequences to retain fragments of the genomes of viruses that the bacteria have come into contact with in the past, which allows them to block the virus's replication through a form of RNA interference.[23][24] This genetic system provides bacteria with acquired immunity to infection.[25]

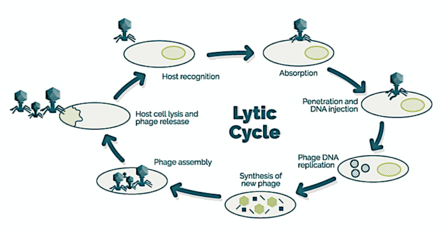
→ attachment: the phage attaches itself to the surface of the host cell
→ penetration: the phage injects its DNA through the cell membrane
→ transcription: the host cell's DNA is degraded and the cell's metabolism is directed to initiate phage biosynthesis
→ biosynthesis: the phage DNA replicates inside the cell
→ maturation: the replicated material assembles into fully formed viral phages
→ lysis: the newly formed phages are released from the infected cell (which is itself destroyed in the process) to seek out new host cells [26]
Microbes drive the nutrient transformations that sustain Earth's ecosystems,[27] and the viruses that infect these microbes modulate both microbial population size and diversity.[28][21] The cyanobacterium Prochlorococcus, the most abundant oxygenic phototroph on Earth, contributes a substantial fraction of global primary carbon production, and often reaches densities of over 100,000 cells per milliliter in oligotrophic and temperate oceans.[29] Hence, viral (cyanophage) infection and lysis of Prochlorococcus represent an important component of the global carbon cycle. In addition to their ecological role in inducing host mortality, cyanophages influence the metabolism and evolution of their hosts by co-opting and exchanging genes, including core photosynthesis genes.[21]

For a long time, tailed phages of the order Caudovirales seemed to dominate marine ecosystems in number and diversity of organisms.[17] However, as a result of more recent research, non-tailed viruses appear to dominate multiple depths and oceanic regions.[31] These non-tailed phages also infect marine bacteria, and include the families Corticoviridae,[32] Inoviridae,[33] Microviridae [34] and Autolykiviridae.[35][36][37][38]
As of September 2023, Halomonas phage vB HmeY H4907 is the first virus isolated from the deepest part of the ocean.[39]
Archaeal viruses
[edit]
Archaean viruses replicate within archaea: these are double-stranded DNA viruses with unusual and sometimes unique shapes.[40][41] These viruses have been studied in most detail in the thermophilic archaea, particularly the orders Sulfolobales and Thermoproteales.[42] Defences against these viruses involve RNA interference from repetitive DNA sequences within archaean genomes that are related to the genes of the viruses.[43][44] Most archaea have CRISPR–Cas systems as an adaptive defence against viruses. These enable archaea to retain sections of viral DNA, which are then used to target and eliminate subsequent infections by the virus using a process similar to RNA interference.[45]
Fungal viruses
[edit]Mycoviruses, also known as mycophages, are viruses that infect fungi. The infection of fungal cells is different from that of animal cells. Fungi have a rigid cell wall made of chitin, so most viruses can get inside these cells only after trauma to the cell wall.[46]
- See, Nerva L, Ciuffo M, Vallino M, Margaria P, Varese G, Gnavi G, Turina M (2016). "Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi". Virus Research. 219: 22–38. doi:10.1016/j.virusres.2015.10.028. hdl:2318/1527617. PMID 26546154. S2CID 53417720.
Eukaryote viruses
[edit]
Marine protists
[edit]By 2015, about 40 viruses affecting marine protists had been isolated and examined, most of them viruses of microalgae.[47] The genomes of these marine protist viruses are highly diverse.[48][49] Marine algae can be infected by viruses in the family Phycodnaviridae. These are large (100–560 kb) double-stranded DNA viruses with icosahedral shaped capsids. By 2014, 33 species divided into six genera had been identified within the family,[50][51] which belongs to a super-group of large viruses known as nucleocytoplasmic large DNA viruses. Evidence was published in 2014 suggesting some strains of Phycodnaviridae might infect humans rather than just algal species, as was previously believed.[52] Most genera under this family enter the host cell by cell receptor endocytosis and replicate in the nucleus.

Phycodnaviridae play important ecological roles by regulating the growth and productivity of their algal hosts. Algal species such Heterosigma akashiwo and the genus Chrysochromulina can form dense blooms which can be damaging to fisheries, resulting in losses in the aquaculture industry.[53] Heterosigma akashiwo virus (HaV) has been suggested for use as a microbial agent to prevent the recurrence of toxic red tides produced by this algal species.[54] The coccolithovirus Emiliania huxleyi virus 86, a giant double-stranded DNA virus, infects the ubiquitous coccolithophore Emiliania huxleyi.[50][51] This virus has one of the largest known genomes among marine viruses.[55] Phycodnaviridae cause death and lysis of freshwater and marine algal species, liberating organic carbon, nitrogen and phosphorus into the water, providing nutrients for the microbial loop.[56]
The virus-to-prokaryote ratio, VPR, is often used as an indicator of the relationship between viruses and hosts. Studies have used VPR to indirectly infer virus impact on marine microbial productivity, mortality, and biogeochemical cycling.[57] However, in making these approximations, scientists assume a VPR of 10:1, the median observed VPR in the surface ocean.[57][18] The actual VPR varies greatly depending on location, so VPR may not be the accurate proxy for viral activity or abundance as it has been treated.[57][58]
Marine invertebrates
[edit]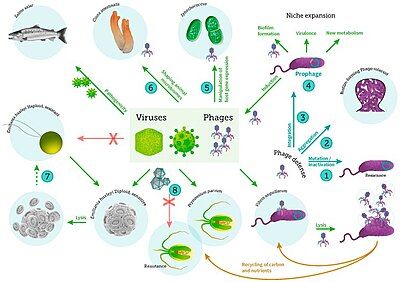
including viral infection of bacteria, phytoplankton and fish[59]
Marine invertebrates are susceptible to viral diseases.[60][61][62] Sea star wasting disease is a disease of starfish and several other echinoderms that appears sporadically, causing mass mortality of those affected.[63] There are around 40 different species of sea stars that have been affected by this disease. In 2014 it was suggested that the disease is associated with a single-stranded DNA virus now known as the sea star-associated densovirus (SSaDV); however, sea star wasting disease is not fully understood.[64]
Marine vertebrates
[edit]Fish are particularly prone to infections with rhabdoviruses, which are distinct from, but related to rabies virus. At least nine types of rhabdovirus cause economically important diseases in species including salmon, pike, perch, sea bass, carp and cod. The symptoms include anaemia, bleeding, lethargy and a mortality rate that is affected by the temperature of the water. In hatcheries the diseases are often controlled by increasing the temperature to 15–18 °C (59–64 °F).[65]: 442–443 Like all vertebrates, fish suffer from herpes viruses. These ancient viruses have co-evolved with their hosts and are highly species-specific.[65]: 324 In fish, they cause cancerous tumours and non-cancerous growths called hyperplasia.[65]: 325
In 1984, infectious salmon anemia (ISAv) was discovered in Norway in an Atlantic salmon hatchery. Eighty per cent of the fish in the outbreak died. ISAv, a viral disease, is now a major threat to the viability of Atlantic salmon farming.[66] As the name implies, it causes severe anemia of infected fish. Unlike mammals, the red blood cells of fish have DNA and can become infected with viruses. Management strategies include developing a vaccine and improving genetic resistance to the disease.[67]
Marine mammals are also susceptible to marine viral infections. In 1988 and 2002, thousands of harbour seals were killed in Europe by phocine distemper virus.[68] Many other viruses, including caliciviruses, herpesviruses, adenoviruses and parvoviruses, circulate in marine mammal populations.[69]
Giant marine viruses
[edit]Most viruses range in length from about 20 to 300 nanometers. This can be contrasted with the length of bacteria, which starts at about 400 nanometers. There are also giant viruses, often called giruses, typically about 1000 nanometers (one micron) in length. All giant viruses belong to the phylum Nucleocytoviricota (NCLDV), together with poxviruses. The largest known of these is Tupanvirus. This genus of giant virus was discovered in 2018 in the deep ocean as well as a soda lake, and can reach up to 2.3 microns in total length.[70]

-
The giant mimivirus
The discovery and subsequent characterization of giant viruses has triggered some debate concerning their evolutionary origins. The two main hypotheses for their origin are that either they evolved from small viruses, picking up DNA from host organisms, or that they evolved from very complicated organisms into the current form which is not self-sufficient for reproduction.[72] What sort of complicated organism giant viruses might have diverged from is also a topic of debate. One proposal is that the origin point actually represents a fourth domain of life,[73][74] but this has been largely discounted.[75][76]
Virophages
[edit]Virophages are small, double-stranded DNA viruses that rely on the co-infection of giant viruses. Virophages rely on the viral replication factory of the co-infecting giant virus for their own replication. One of the characteristics of virophages is that they have a parasitic relationship with the co-infecting virus. Their dependence upon the giant virus for replication often results in the deactivation of the giant viruses. The virophage may improve the recovery and survival of the host organism. Unlike other satellite viruses, virophages have a parasitic effect on their co-infecting virus. Virophages have been observed to render a giant virus inactive and thereby improve the condition of the host organism.
All known virophages are grouped into the family Lavidaviridae (from "large virus dependent or associated" + -viridae).[77] The first virophage was discovered in a cooling tower in Paris in 2008. It was discovered with its co-infecting giant virus, Acanthamoeba castellanii mamavirus (ACMV). The virophage was named Sputnik and its replication relied entirely on the co-infection of ACMV and its cytoplasmic replication machinery. Sputnik was also discovered to have an inhibitory effect on ACMV and improved the survival of the host. Other characterised virophages include Sputnik 2, Sputnik 3, Zamilon and Mavirus.[78][79]
Most of these virophages were discovered by analyzing metagenomic data sets. In metagenomic analysis, DNA sequences are run through multiple bioinformatic algorithms which pull out certain important patterns and characteristics. In these data sets are giant viruses and virophages. They are separated by looking for sequences around 17 to 20 kbp long which have similarities to already sequenced virophages. These virophages can have linear or circular double-stranded DNA genomes.[80] Virophages in culture have icosahedral capsid particles that measure around 40 to 80 nanometers long.[81] Virophage particles are so small that electron microscopy must be used to view these particles. Metagenomic sequence-based analyses have been used to predict around 57 complete and partial virophage genomes[82] and in December 2019 to identify 328 high-quality (complete or near-complete) genomes from diverse habitats including the human gut, plant rhizosphere, and terrestrial subsurface, from 27 distinct taxonomic clades.[83]
A giant marine virus CroV infects and causes the death by lysis of the marine zooflagellate Cafeteria roenbergensis.[85] This impacts coastal ecology because Cafeteria roenbergensis feeds on bacteria found in the water. When there are low numbers of Cafeteria roenbergensis due to extensive CroV infections, the bacterial populations rise exponentially.[86] The impact of CroV on natural populations of C. roenbergensis remains unknown; however, the virus has been found to be very host specific, and does not infect other closely related organisms.[87] Cafeteria roenbergensis is also infected by a second virus, the Mavirus virophage, during co-infection with CroV.[78] This virus interferes with the replication of CroV, which leads to the survival of C. roenbergensis cells. Mavirus is able to integrate into the genome of cells of C. roenbergensis and thereby confer immunity to the population.[79]
Role of marine viruses
[edit]Although marine viruses have only recently been studied extensively, they are already known to hold critical roles in many ecosystem functions and cycles.[88] Marine viruses offer a number of important ecosystem services and are essential to the regulation of marine ecosystems.[3] Marine bacteriophages and other viruses appear to influence biogeochemical cycles globally, provide and regulate microbial biodiversity, cycle carbon through marine food webs, and are essential in preventing bacterial population explosions.[89]
Viral shunt
[edit]
The dominant hosts for viruses in the ocean are marine microorganisms, such as bacteria.[14] Bacteriophages are harmless to plants and animals, and are essential to the regulation of marine and freshwater ecosystems[91] are important mortality agents of phytoplankton, the base of the foodchain in aquatic environments.[92] They infect and destroy bacteria in aquatic microbial communities, and are one of the most important mechanisms of recycling carbon and nutrient cycling in marine environments. The organic molecules released from the dead bacterial cells stimulate fresh bacterial and algal growth, in a process known as the viral shunt.[93]
In this way, marine viruses are thought to play an important role in nutrient cycles by increasing the efficiency of the biological pump. Viruses cause lysis of living cells, that is, they break the cell membranes down. This releases compounds such as amino acids and nucleic acids, which tend to be recycled near the surface.
Viral activity also enhances the ability of the biological pump to sequester carbon in the deep ocean.[69] Lysis releases more indigestible carbon-rich material like that found in cell walls, which is likely exported to deeper waters. Thus, the material that is exported to deeper waters by the viral shunt is probably more carbon rich than the material from which it was derived.[94][95] By increasing the amount of respiration in the oceans, viruses are indirectly responsible for reducing the amount of carbon dioxide in the atmosphere by about three gigatonnes of carbon per year.[69] Lysis of bacteria by viruses has been shown to also enhance nitrogen cycling and stimulate phytoplankton growth.[96]
The viral shunt pathway is a mechanism that prevents (prokaryotic and eukaryotic) marine microbial particulate organic matter (POM) from migrating up trophic levels by recycling them into dissolved organic matter (DOM), which can be readily taken up by microorganisms. Viral shunting helps maintain diversity within the microbial ecosystem by preventing a single species of marine microbe from dominating the micro-environment.[97] The DOM recycled by the viral shunt pathway is comparable to the amount generated by the other main sources of marine DOM.[98]


Viruses are the most abundant biological entity in marine environments.[5] On average there are about ten million of them in one milliliter of seawater.[100] Most of these viruses are bacteriophages infecting heterotrophic bacteria and cyanophages infecting cyanobacteria. Viruses easily infect microorganisms in the microbial loop due to their relative abundance compared to microbes.[101] Prokaryotic and eukaryotic mortality contribute to carbon nutrient recycling through cell lysis. There is evidence as well of nitrogen (specifically ammonium) regeneration. This nutrient recycling helps stimulates microbial growth.[102] As much as 25% of the primary production from phytoplankton in the global oceans may be recycled within the microbial loop through viral shunting.[103]
Regulating the carbon cycle
[edit]Viruses act as "regulators" of the global carbon cycle because they impact the material cycles and energy flows of food webs and the microbial loop. The average contribution of viruses to the Earth ecosystem carbon cycle is 8.6%, of which its contribution to marine ecosystems (1.4%) is less than its contribution to terrestrial (6.7%) and freshwater (17.8%) ecosystems. Over the past 2,000 years, anthropogenic activities and climate change have gradually altered the regulatory role of viruses in ecosystem carbon cycling processes. This has been particularly conspicuous over the past 200 years due to rapid industrialization and the attendant population growth.[104]

Limiting algal blooms
[edit]Microorganisms make up about 70% of the marine biomass.[105] It is estimated viruses kill 20% of the microorganism biomass each day and that there are 15 times as many viruses in the oceans as there are bacteria and archaea. Viruses are the main agents responsible for the rapid destruction of harmful algal blooms,[106] which often kill other marine life.[107] Scientists are exploring the potential of marine cyanophages to be used to prevent or reverse eutrophication. The number of viruses in the oceans decreases further offshore and deeper into the water, where there are fewer host organisms.[108]
Gene transfer
[edit]Marine bacteriophages often contain auxiliary metabolic genes, host-derived genes thought to sustain viral replication by supplementing host metabolism during viral infection.[109] These genes can impact multiple biogeochemical cycles, including carbon, phosphorus, sulfur, and nitrogen.[110][111][112][113]
Viruses are an important natural means of transferring genes between different species, which increases genetic diversity and drives evolution.[10] It is thought viruses played a central role in early evolution, before bacteria, archaea and eukaryotes diversified, at the time of the last universal common ancestor of life on Earth.[114] Viruses are still one of the largest reservoirs of unexplored genetic diversity on Earth.[108]
Marine habitats
[edit]Along the coast
[edit]Marine coastal habitats sit at the interface between the land and the ocean. It is likely that RNA viruses play significant roles in these environments.[115]
At the ocean surface
[edit]
Marine surface habitats sit at the interface between the atmosphere and the ocean. The biofilm-like habitat at the surface of the ocean harbours surface-dwelling microorganisms, commonly referred to as neuston. Viruses in the microlayer, the so-called virioneuston, have recently become of interest to researchers as enigmatic biological entities in the boundary surface layers with potentially important ecological impacts. Given this vast air–water interface sits at the intersection of major air–water exchange processes spanning more than 70% of the global surface area, it is likely to have profound implications for marine biogeochemical cycles, on the microbial loop and gas exchange, as well as the marine food web structure, the global dispersal of airborne viruses originating from the sea surface microlayer, and human health.[116]
In the water column
[edit]Marine viral activity presents a potential explanation of the paradox of the plankton proposed by George Hutchinson in 1961.[117] The paradox of the plankton is that many plankton species have been identified in small regions in the ocean where limited resources should create competitive exclusion, limiting the number of coexisting species.[117] Marine viruses could play a role in this effect, as viral infection increases as potential contact with hosts increases.[4] Viruses could therefore control the populations of plankton species that grow too abundant, allowing a wide diversity of species to coexist.[4]
In sediments
[edit]Marine bacteriophages play an important role in deep sea ecosystems. There are between 5x1012 and 1x1013 phages per square metre in deep sea sediments and their abundance closely correlates with the number of prokaryotes found in the sediments. They are responsible for the death of 80% of the prokaryotes found in the sediments, and almost all of these deaths are caused by cell lysis (bursting). This allows nitrogen, carbon, and phosphorus from the living cells to be converted into dissolved organic matter and detritus, contributing to the high rate of nutrient turnover in deep sea sediments. Because of the importance of deep sea sediments in biogeochemical cycles, marine bacteriophages influence the carbon, nitrogen and phosphorus cycles. More research needs to be done to more precisely elucidate these influences.[118]
In hydrothermal vents
[edit]Viruses are part of the hydrothermal vent microbial community and their influence on the microbial ecology in these ecosystems is a burgeoning field of research.[119] Viruses are the most abundant life in the ocean, harboring the greatest reservoir of genetic diversity.[106] As their infections are often fatal, they constitute a significant source of mortality and thus have widespread influence on biological oceanographic processes, evolution and biogeochemical cycling within the ocean.[108] Evidence has been found however to indicate that viruses found in vent habitats have adopted a more mutualistic than parasitic evolutionary strategy in order to survive the extreme and volatile environment they exist in.[120] Deep-sea hydrothermal vents were found to have high numbers of viruses indicating high viral production.[121] Like in other marine environments, deep-sea hydrothermal viruses affect abundance and diversity of prokaryotes and therefore impact microbial biogeochemical cycling by lysing their hosts to replicate.[122] However, in contrast to their role as a source of mortality and population control, viruses have also been postulated to enhance survival of prokaryotes in extreme environments, acting as reservoirs of genetic information. The interactions of the virosphere with microorganisms under environmental stresses is therefore thought to aide microorganism survival through dispersal of host genes through horizontal gene transfer.[123]
Polar regions
[edit]In addition to varied topographies and in spite of an extremely cold climate, the polar aquatic regions are teeming with microbial life. Even in sub-glacial regions, cellular life has adapted to these extreme environments where perhaps there are traces of early microbes on Earth. As grazing by macrofauna is limited in most of these polar regions, viruses are being recognised for their role as important agents of mortality, thereby influencing the biogeochemical cycling of nutrients that, in turn, impact community dynamics at seasonal and spatial scales.[124] The polar regions are characterised by truncated food webs, and the role of viruses in ecosystem function is likely to be even greater than elsewhere in the marine food web, yet their diversity is still relatively under-explored, and the way in which they affect polar communities is not well understood,[125] particularly in nutrient cycling.[126][127][128][124]
Distribution
[edit]Viruses are highly host specific.[129] A marine virus is more likely to infect cooccurring organisms, those that live in the same region the virus lives in.[130] Therefore, biogeography is an important factor in a virion's ability to infect.
Knowledge of this variation in viral populations across spatiotemporal and other environmental gradients is supported by viral morphology, as determined by transmission electron microscopy (TEM). Non-tailed viruses appear to be dominant in multiple depths and oceanic regions, followed by the Caudovirales myoviruses, podoviruses, and siphoviruses.[31] However, viruses belonging to families Corticoviridae,[131] Inoviridae[132] and Microviridae[133] are also known to infect diverse marine bacteria. Metagenomic evidence suggests that microviruses (icosahedral ssDNA phages) are particularly prevalent in marine habitats.[133]
Metagenomic approaches to assess viral diversity are often limited by a lack of reference sequences, leaving many sequences unannotated.[134] However, viral contigs are generated through direct sequencing of a viral fraction, typically generated after 0.02-um filtration of a marine water sample, or through bioinformatics approaches to identify viral contigs or viral genomes from a microbial metagenome. Novel tools to identify putative viral contigs, such as VirSorter[135] and VirFinder,[136] allow for the assessment of patterns of viral abundance, host range, and functional content of marine bacteriophage.[137][138]
See also
[edit]References
[edit]- ^ Bonnain C, Breitbart M, Buck K (2016). "The ferrojan horse hypothesis: iron-virus interactions in the ocean". Frontiers in Marine Science. 3: 82. doi:10.3389/fmars.2016.00082. S2CID 2917222.
- ^ These Tiny Organisms Have Some Really Weird Shapes, National Geographic, 12 November 2016.
- ^ a b Shors T (2008). Understanding Viruses. Jones and Bartlett Publishers. p. 5. ISBN 978-0-7637-2932-5.
- ^ a b c d Brussaard CP, Baudoux AC, Rodríguez-Valera F (2016). Stal LJ, Cretoiu MS (eds.). Marine Viruses. Springer International Publishing. pp. 155–183. doi:10.1007/978-3-319-33000-6_5. ISBN 9783319329987.
{{cite book}}:|work=ignored (help) - ^ a b c d Koonin EV, Senkevich TG, Dolja VV (2006). "The ancient Virus World and evolution of cells". Biology Direct. 1: 29. doi:10.1186/1745-6150-1-29. PMC 1594570. PMID 16984643.
{{cite journal}}: CS1 maint: multiple names: authors list (link) Modified text was copied from this source, which is available under a Creative Commons Attribution 2.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 2.0 International License.
- ^ Suttle C (2005). "Viruses in the sea". Nature. 437 (7057): 356–361. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- ^ Mahy WJ, Van Regenmortel MHV, eds. (2009). Desk Encyclopedia of General Virology. Oxford: Academic Press. p. 28. ISBN 978-0-12-375146-1.
- ^ Iyer LM, Balaji S, Koonin EV, Aravind L (2006). "Evolutionary genomics of nucleo-cytoplasmic large DNA viruses". Virus Research. 117 (1): 156–84. doi:10.1016/j.virusres.2006.01.009. PMID 16494962.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R (October 2010). "Viral mutation rates". Journal of Virology. 84 (19): 9733–48. doi:10.1128/JVI.00694-10. PMC 2937809. PMID 20660197.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H (2003). "Phage as agents of lateral gene transfer". Current Opinion in Microbiology. 6 (4): 417–24. doi:10.1016/S1369-5274(03)00086-9. PMID 12941415.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Koonin EV, Starokadomskyy P (2016). "Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question". Studies in History and Philosophy of Biological and Biomedical Sciences. 59: 125–134. doi:10.1016/j.shpsc.2016.02.016. PMC 5406846. PMID 26965225.
- ^ Koonin EV, Starokadomskyy P (7 March 2016). "Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question". Studies in History and Philosophy of Biological and Biomedical Sciences. 59: 125–34. doi:10.1016/j.shpsc.2016.02.016. PMC 5406846. PMID 26965225.
- ^ Rybicki, EP (1990). "The classification of organisms at the edge of life, or problems with virus systematics". South African Journal of Science. 86: 182–186.
- ^ a b Mann NH (May 2005). "The third age of phage". PLOS Biology. 3 (5): e182. doi:10.1371/journal.pbio.0030182. PMC 1110918. PMID 15884981.
- ^ Wommack KE, Hill RT, Muller TA, Colwell RR (April 1996). "Effects of sunlight on bacteriophage viability and structure". Applied and Environmental Microbiology. 62 (4): 1336–41. Bibcode:1996ApEnM..62.1336W. doi:10.1128/AEM.62.4.1336-1341.1996. PMC 167899. PMID 8919794.
- ^ Marie D, Brussaard CP, Thyrhaug R, Bratbak G, Vaulot D (January 1999). "Enumeration of marine viruses in culture and natural samples by flow cytometry". Applied and Environmental Microbiology. 65 (1): 45–52. Bibcode:1999ApEnM..65...45M. doi:10.1128/AEM.65.1.45-52.1999. PMC 90981. PMID 9872758.
- ^ a b Mann NH (17 May 2005). "The Third Age of Phage". PLOS Biology. 3 (5): 753–755. doi:10.1371/journal.pbio.0030182. PMC 1110918. PMID 15884981.
- ^ a b Wommack KE, Colwell RR (March 2000). "Virioplankton: viruses in aquatic ecosystems". Microbiology and Molecular Biology Reviews. 64 (1): 69–114. doi:10.1128/MMBR.64.1.69-114.2000. PMC 98987. PMID 10704475.
- ^ Bergh O, Børsheim KY, Bratbak G, Heldal M (August 1989). "High abundance of viruses found in aquatic environments". Nature. 340 (6233): 467–68. Bibcode:1989Natur.340..467B. doi:10.1038/340467a0. PMID 2755508. S2CID 4271861.
- ^ Shors pp. 595–97
- ^ a b c d Murata K, Zhang Q, Galaz-Montoya J, Fu C, Coleman M, Osburne M, Schmid M, Sullivan M, Chisholm S, Chiu W (2017). "Visualizing adsorption of cyanophage P-SSP7 onto marine Prochlorococcus". Scientific Reports. 7: 44176. Bibcode:2017NatSR...744176M. doi:10.1038/srep44176. PMC 5345008. PMID 28281671.
- ^ Bickle TA, Krüger DH (June 1993). "Biology of DNA restriction". Microbiological Reviews. 57 (2): 434–50. doi:10.1128/MMBR.57.2.434-450.1993. PMC 372918. PMID 8336674.
- ^ Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (March 2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science. 315 (5819): 1709–12. Bibcode:2007Sci...315.1709B. doi:10.1126/science.1138140. hdl:20.500.11794/38902. PMID 17379808. S2CID 3888761.
- ^ Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (August 2008). "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science. 321 (5891): 960–64. Bibcode:2008Sci...321..960B. doi:10.1126/science.1159689. PMC 5898235. PMID 18703739.
- ^ Mojica FJ, Rodriguez-Valera F (September 2016). "The discovery of CRISPR in archaea and bacteria". The FEBS Journal. 283 (17): 3162–69. doi:10.1111/febs.13766. hdl:10045/57676. PMID 27234458. S2CID 42827598.
- ^ How do bacteriophages reproduce? University of Barcelona. Retrieved 12 July 2020.
- ^ Falkowski P, Fenchel T, Delong E (2008). "The microbial engines that drive Earth's biogeochemical cycles". Science. 320 (5879): 1034–1039. Bibcode:2008Sci...320.1034F. doi:10.1126/science.1153213. PMID 18497287. S2CID 2844984.
- ^ Brum J, Sullivan M (2015). "Rising to the challenge: accelerated pace of discovery transforms marine virology". Nature Reviews Microbiology. 13 (3): 147–159. doi:10.1038/nrmicro3404. PMID 25639680. S2CID 32998525.
- ^ Bouman H, Ulloa O, Scanlan D, Zwirglmaier K, Li W, Platt T, Stuart V, Barlow R, Leth O, Clementson L, Lutz V (2006). "Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes". Science. 312 (5775): 918–921. Bibcode:2006Sci...312..918B. doi:10.1126/science.1122692. PMID 16690867. S2CID 20738145.
- ^ Gonzalez-Serrano R, Dunne M, Rosselli R, Martin-Cuadrado AB, Grosboillot V, Zinsli LV, Roda-Garcia JJ, Loessner MJ, Rodriguez-Valera F (2020). "Alteromonas Myovirus V22 Represents a New Genus of Marine Bacteriophages Requiring a Tail Fiber Chaperone for Host Recognition". mSystems. 5 (3). doi:10.1128/mSystems.00217-20. PMC 7289586. PMID 32518192.
- ^ a b Brum JR, Schenck RO, Sullivan MB (September 2013). "Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses". The ISME Journal. 7 (9): 1738–51. Bibcode:2013ISMEJ...7.1738B. doi:10.1038/ismej.2013.67. PMC 3749506. PMID 23635867.
- ^ Krupovic M, Bamford DH (2007). "Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria". BMC Genomics. 8: 236. doi:10.1186/1471-2164-8-236. PMC 1950889. PMID 17634101.
- ^ Xue H, Xu Y, Boucher Y, Polz MF (2012). "High Frequency of a Novel Filamentous Phage, VCYϕ, within an Environmental Vibrio cholerae Population". Applied and Environmental Microbiology. 78 (1): 28–33. Bibcode:2012ApEnM..78...28X. doi:10.1128/AEM.06297-11. PMC 3255608. PMID 22020507.
- ^ Roux S, Krupovic M, Poulet A, Debroas D, Enault F (2012). "Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads". PLOS ONE. 7 (7): e40418. Bibcode:2012PLoSO...740418R. doi:10.1371/journal.pone.0040418. PMC 3394797. PMID 22808158.
- ^ Kauffman KM, Hussain FA, Yang J, Arevalo P, Brown JM, Chang WK, Vaninsberghe D, Elsherbini J, Sharma RS, Cutler MB, Kelly L, Polz MF (2018). "A major lineage of non-tailed dsDNA viruses as unrecognized killers of marine bacteria". Nature. 554 (7690): 118–122. Bibcode:2018Natur.554..118K. doi:10.1038/nature25474. PMID 29364876. S2CID 4462007.
- ^ Scientists Find New Type of Virus in World's Oceans: Autolykiviridae, on: sci-news, 25 January 2018
- ^ Never-Before-Seen Viruses With Weird DNA Were Just Discovered in The Ocean, on: sciencealert, 25 January 2018
- ^ NCBI: Autolykiviridae (family) – unclassified dsDNA viruses
- ^ Su Y, Zhang W, Liang Y, Wang H, Liu Y, Zheng K, Liu Z, Yu H, Ren L, Shao H, Sung YY, Mok WJ, Wong LL, Zhang YZ, McMinn A (20 September 2023). Chao DY (ed.). "Identification and genomic analysis of temperate Halomonas bacteriophage vB_HmeY_H4907 from the surface sediment of the Mariana Trench at a depth of 8,900 m". Microbiology Spectrum. 11 (5): e0191223. doi:10.1128/spectrum.01912-23. ISSN 2165-0497. PMC 10580944. PMID 37728551.
- ^ Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ (2009). "Structural and functional studies of archaeal viruses". Journal of Biological Chemistry. 284 (19): 12599–603. doi:10.1074/jbc.R800078200. PMC 2675988. PMID 19158076.
- ^ Prangishvili D, Forterre P, Garrett RA (2006). "Viruses of the Archaea: a unifying view". Nature Reviews Microbiology. 4 (11): 837–48. doi:10.1038/nrmicro1527. PMID 17041631. S2CID 9915859.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Prangishvili D, Garrett RA (2004). "Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses" (PDF). Biochemical Society Transactions. 32 (Pt 2): 204–8. doi:10.1042/BST0320204. PMID 15046572. S2CID 20018642.
- ^ Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E (February 2005). "Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements". Journal of Molecular Evolution. 60 (2): 174–82. Bibcode:2005JMolE..60..174M. doi:10.1007/s00239-004-0046-3. PMID 15791728. S2CID 27481111.
- ^ Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (March 2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biology Direct. 1: 7. doi:10.1186/1745-6150-1-7. PMC 1462988. PMID 16545108.
- ^ van der Oost J, Westra ER, Jackson RN, Wiedenheft B (July 2014). "Unravelling the structural and mechanistic basis of CRISPR-Cas systems". Nature Reviews Microbiology. 12 (7): 479–92. doi:10.1038/nrmicro3279. PMC 4225775. PMID 24909109.
- ^ Dimmock NJ, Easton AJ, Leppard K (2007). Introduction to Modern Virology (Sixth ed.). Blackwell Publishing. p. 70. ISBN 978-1-4051-3645-7.
- ^ Tomaru Y, Kimura K and Nagasaki K (2015) "Marine Protist Viruses". In: Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, Not F (eds) Marine Protists pages 501–517. Springer, Tokyo. doi:10.1007/978-4-431-55130-0_20. ISBN 978-4-431-55130-0.
- ^ Hyman P, Abedon ST (2012). "Smaller Fleas: Viruses of Microorganisms". Scientifica. 2012: 1–23. doi:10.6064/2012/734023. PMC 3820453. PMID 24278736..
 Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
- ^ Short S (2012). "The ecology of viruses that infect eukaryotic algae". Environmental Microbiology. 14 (9): 2253–2271. Bibcode:2012EnvMi..14.2253S. doi:10.1111/j.1462-2920.2012.02706.x. PMID 22360532.
- ^ a b "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ^ a b ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ^ Yolken RH, et al. (2014). "Chlorovirus ATCV-1 is part of the human oropharyngeal virome and is associated with changes in cognitive functions in humans and mice". Proc Natl Acad Sci U S A. 111 (45): 16106–16111. Bibcode:2014PNAS..11116106Y. doi:10.1073/pnas.1418895111. PMC 4234575. PMID 25349393.
- ^ Tomaru Y, Shirai Y, Nagasaki K (1 August 2008). "Ecology, physiology and genetics of a phycodnavirus infecting the noxious bloom-forming raphidophyte Heterosigma akashiwo". Fisheries Science. 74 (4): 701–711. Bibcode:2008FisSc..74..701T. doi:10.1111/j.1444-2906.2008.01580.x. S2CID 23152411.
- ^ Nagasaki K, Tarutani K, Yamaguchi M (1 March 1999). "Growth Characteristics of Heterosigma akashiwo Virus and Its Possible Use as a Microbiological Agent for Red Tide Control". Applied and Environmental Microbiology. 65 (3): 898–902. Bibcode:1999ApEnM..65..898N. doi:10.1128/AEM.65.3.898-902.1999. PMC 91120. PMID 10049839.
- ^ Largest known viral genomes Giantviruses.org. Accessed: 11 June 2020.
- ^ Sigee D (27 September 2005). Freshwater Microbiology: Biodiversity and Dynamic Interactions of Microorganisms in the Aquatic Environment. John Wiley & Sons. ISBN 9780470026472.
- ^ a b c Wigington CH, Sonderegger DL, Brussaard CP, Buchan A, Finke JF, Fuhrman J, Lennon JT, Middelboe M, Suttle CA, Stock C, Wilson WH (26 August 2015). "Re-examining the relationship between virus and microbial cell abundances in the global oceans". bioRxiv: 025544. bioRxiv 10.1101/025544. doi:10.1101/025544.
- ^ Parikka KJ, Le Romancer M, Wauters N, Jacquet S (May 2017). "Deciphering the virus-to-prokaryote ratio (VPR): insights into virus-host relationships in a variety of ecosystems". Biological Reviews of the Cambridge Philosophical Society. 92 (2): 1081–1100. doi:10.1111/brv.12271. PMID 27113012. S2CID 3463306.
- ^ Middelboe M, Brussaard C (2017). "Marine viruses: key players in marine ecosystems". Viruses. 9 (10): 302. doi:10.3390/v9100302. PMC 5691653. PMID 29057790.
- ^ TJohnson P (1984). "Viral diseases of marine invertebrates". Helgoländer Meeresuntersuchungen. 37 (1–4): 65–98. Bibcode:1984HM.....37...65J. doi:10.1007/BF01989296. S2CID 30161955.
- ^ Renault T (2011) "Viruses infecting marine molluscs" In: Hurst CJ (Ed) Studies in Viral Ecology, Volume 2: Animal Host Systems, John Wiley & Sons. ISBN 9781118024584.
- ^ Arzul I, Corbeil S, Morga B, Renault T (2017). "Viruses infecting marine molluscs" (PDF). Journal of Invertebrate Pathology. 147: 118–135. Bibcode:2017JInvP.147..118A. doi:10.1016/j.jip.2017.01.009. PMID 28189502.
- ^ Dawsoni, Solaster. "Sea Star Species Affected by Wasting Syndrome." Pacificrockyintertidal.org Seastarwasting.org (n.d.): n. pag. Ecology and Evolutionary Biology. Web.
- ^ "Sea Star Wasting Syndrome | MARINe". eeb.ucsc.edu. Retrieved 3 June 2018.
- ^ a b c Murphy, FA, Gibbs, EPJ, Horzinek, MC, Studdart MJ (1999). Veterinary Virology. Boston: Academic Press. ISBN 978-0-12-511340-3.
- ^ New Brunswick to help Chile beat disease Fish Information and Services
- ^ Fact Sheet - Atlantic Salmon Aquaculture Research Archived December 29, 2010, at the Wayback Machine Fisheries and Oceans Canada. Retrieved 12 May 2009.
- ^ Hall AJ, Jepson PD, Goodman SJ, Harkonen T (2006). "Phocine distemper virus in the North and European Seas – data and models, nature and nurture". Biological Conservation. 131 (2): 221–29. Bibcode:2006BCons.131..221H. doi:10.1016/j.biocon.2006.04.008.
- ^ a b c Suttle CA (October 2007). "Marine viruses – major players in the global ecosystem". Nature Reviews Microbiology. 5 (10): 801–12. doi:10.1038/nrmicro1750. PMID 17853907. S2CID 4658457.
- ^ Abrahão J, Silva L, Silva LS, Khalil JY, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, Scola BL (27 February 2018). "Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere". Nature Communications. 9 (1): 749. Bibcode:2018NatCo...9..749A. doi:10.1038/s41467-018-03168-1. PMC 5829246. PMID 29487281.
- ^ Xiao C, Fischer M, Bolotaulo D, Ulloa-Rondeau N, Avila G, Suttle C (2017). "Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses". Scientific Reports. 7 (1): 5484. Bibcode:2017NatSR...7.5484X. doi:10.1038/s41598-017-05824-w. PMC 5511168. PMID 28710447.
- ^ Bichell RE. "In Giant Virus Genes, Hints About Their Mysterious Origin". All Things Considered.
- ^ Van Etten JL (July–August 2011). "Giant Viruses". American Scientist. 99 (4): 304–311. doi:10.1511/2011.91.304.
- ^ Legendre M, Arslan D, Abergel C, Claverie JM (January 2012). "Genomics of Megavirus and the elusive fourth domain of Life". Communicative & Integrative Biology. 5 (1): 102–6. doi:10.4161/cib.18624. PMC 3291303. PMID 22482024.
- ^ Schulz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, Kyrpides NC, Koonin EV, Woyke T (April 2017). "Giant viruses with an expanded complement of translation system components" (PDF). Science. 356 (6333): 82–85. Bibcode:2017Sci...356...82S. doi:10.1126/science.aal4657. PMID 28386012. S2CID 206655792.
- ^ Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJ (March 2019). "Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism". mBio. 10 (2): e02497-02418. doi:10.1128/mBio.02497-18. PMC 6401483. PMID 30837339.
- ^ Duponchel S, Fischer MG (March 2019). "Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses". PLOS Pathogens. 15 (3): e1007592. doi:10.1371/journal.ppat.1007592. PMC 6428243. PMID 30897185.
- ^ a b Fischer MG, Suttle CA (April 2011). "A virophage at the origin of large DNA transposons". Science. 332 (6026): 231–4. Bibcode:2011Sci...332..231F. doi:10.1126/science.1199412. PMID 21385722. S2CID 206530677.
- ^ a b Fischer MG, Hackl (December 2016). "Host genome integration and giant virus-induced reactivation of the virophage mavirus". Nature. 540 (7632): 288–91. Bibcode:2016Natur.540..288F. doi:10.1038/nature20593. PMID 27929021. S2CID 4458402.
- ^ Katzourakis A, Aswad A (2014). "The origins of giant viruses, virophages and their relatives in host genomes". BMC Biology. 12: 2–3. doi:10.1186/s12915-014-0051-y. PMC 4096385. PMID 25184667.
- ^ Krupovic M, Kuhn J, Fischer M (Fall 2015). "A classification system for virophages and satellite viruses" (PDF). Archives of Virology. 161 (1): 233–247. doi:10.1007/s00705-015-2622-9. PMID 26446887. S2CID 14196910 – via Springer.
- ^ Roux S, Chan LK, Egan R, Malmstrom RR, McMahon KD, Sullivan MB (2017). "Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics". Nature Communications. 8 (1): 858. Bibcode:2017NatCo...8..858R. doi:10.1038/s41467-017-01086-2. ISSN 2041-1723. PMC 5636890. PMID 29021524.
- ^ Paez-Espino D, Zhou J, Roux S, Nayfach S, Pavlopoulos GA, Schulz F, McMahon KD, Walsh D, Woyke T, Ivanova NN, Eloe-Fadrosh EA, Tringe SG, Kyrpides NC (10 December 2019). "Diversity, evolution, and classification of virophages uncovered through global metagenomics". Microbiome. 7 (1): 157. doi:10.1186/s40168-019-0768-5. PMC 6905037. PMID 31823797.
- ^ Duponchel S, Fischer MG (2019). "Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses". PLOS Pathogens. 15 (3): e1007592. doi:10.1371/journal.ppat.1007592. PMC 6428243. PMID 30897185..
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Fischer, M. G., Allen, M. J., Wilson, W. H., Suttle, C. A. (2010). "Giant virus with a remarkable complement of genes infects marine zooplankton" (PDF). Proceedings of the National Academy of Sciences. 107 (45): 19508–19513. Bibcode:2010PNAS..10719508F. doi:10.1073/pnas.1007615107. PMC 2984142. PMID 20974979.
- ^ Matthias G. Fischer, Michael J. Allen, William H. Wilson, Curtis A. Suttle (2010). "Giant virus with a remarkable complement of genes infects marine zooplankton" (PDF). Proceedings of the National Academy of Sciences. 107 (45): 19508–19513. Bibcode:2010PNAS..10719508F. doi:10.1073/pnas.1007615107. PMC 2984142. PMID 20974979.
- ^ Massana, Ramon, Javier Del Campo, Christian Dinter, Ruben Sommaruga (2007). "Crash of a population of the marine heterotrophic flagellate Cafeteria roenbergensis by viral infection". Environmental Microbiology. 9 (11): 2660–2669. Bibcode:2007EnvMi...9.2660M. doi:10.1111/j.1462-2920.2007.01378.x. PMID 17922751. S2CID 30191542.
- ^ Herndl G, Briand F, eds. (2003). Ecology of marine viruses. CIESM Workshop Monograph 21 [1]
- ^ Waldor MK, Friedman DI, Adhya SL, eds. (2005). Phages: their role in bacterial pathogenesis and biotechnology. Washington DC: ASM Press. pp. 450. ISBN 978-1-55581-307-9.
- ^ Käse L, Geuer JK (2018). "Phytoplankton Responses to Marine Climate Change – an Introduction". YOUMARES 8 – Oceans Across Boundaries: Learning from each other. pp. 55–71. doi:10.1007/978-3-319-93284-2_5. ISBN 978-3-319-93283-5. S2CID 134263396.
- ^ Weitz JS, Wilhelm SW (2013). "An ocean of viruses". The Scientist. 27 (7): 35–39.
- ^ Suttle CA (September 2005). "Viruses in the sea". Nature. 437 (7057): 356–61. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- ^ Wilhelm SW, Suttle CA (1999). "Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs". BioScience. 49 (10): 781–88. doi:10.2307/1313569. JSTOR 1313569.
- ^ Suttle CA (October 2007). "Marine viruses—major players in the global ecosystem". Nature Reviews Microbiology. 5 (10): 801–12. doi:10.1038/nrmicro1750. PMID 17853907. S2CID 4658457.
- ^ Suttle CA (September 2005). "Viruses in the sea". Nature. 437 (7057): 356–61. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- ^ Shelford EJ, Suttle CA (2018). "Virus-mediated transfer of nitrogen from heterotrophic bacteria to phytoplankton". Biogeosciences. 15 (3): 809–15. Bibcode:2018BGeo...15..809S. doi:10.5194/bg-15-809-2018.
- ^ Weinbauer MG, et al. (2007). "Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity". Environmental Microbiology. 9 (3): 777–788. Bibcode:2007EnvMi...9..777W. doi:10.1111/j.1462-2920.2006.01200.x. PMID 17298376.
- ^ Robinson, Carol, and Nagappa Ramaiah. "Microbial heterotrophic metabolic rates constrain the microbial carbon pump." The American Association for the Advancement of Science, 2011.
- ^ Heinrichs, M.E., Mori, C. and Dlugosch, L. (2020) "Complex Interactions Between Aquatic Organisms and Their Chemical Environment Elucidated from Different Perspectives". In: YOUMARES 9-The Oceans: Our Research, Our Future , pages 279–297. Springer. doi:10.1007/978-3-030-20389-4_15.
- ^ Dávila-Ramos S, Castelán-Sánchez HG, Martínez-Ávila L, Sánchez-Carbente MD, Peralta R, Hernández-Mendoza A, Dobson AD, Gonzalez RA, Pastor N, Batista-García RA (2019). "A Review on Viral Metagenomics in Extreme Environments". Frontiers in Microbiology. 10: 2403. doi:10.3389/fmicb.2019.02403. PMC 6842933. PMID 31749771.
- ^ Fuhrman JA (1999). "Marine viruses and their biogeochemical and ecological effects". Nature. 399 (6736): 541–548. Bibcode:1999Natur.399..541F. doi:10.1038/21119. ISSN 0028-0836. PMID 10376593. S2CID 1260399.
- ^ Tsai, An-Yi, Gwo-Ching Gong, and Yu-Wen Huang. "Importance of the Viral Shunt in Nitrogen Cycling in Synechococcus Spp. Growth in Subtropical Western Pacific Coastal Waters." Terrestrial, Atmospheric & Oceanic Sciences25.6 (2014).
- ^ Wilhelm SW, Suttle CA (1999). "Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs". BioScience. 49 (10): 781–788. doi:10.2307/1313569. JSTOR 1313569.
- ^ a b Gao Y, Lu Y, Dungait JA, Liu J, Lin S, Jia J, Yu G (29 March 2022). "The "Regulator" Function of Viruses on Ecosystem Carbon Cycling in the Anthropocene". Frontiers in Public Health. 10. Frontiers Media SA. doi:10.3389/fpubh.2022.858615. ISSN 2296-2565. PMC 9001988.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Bar-On YM, Phillips R, Milo R (2018). "The biomass distribution on Earth". PNAS. 115 (25): 6506–6511. Bibcode:2018PNAS..115.6506B. doi:10.1073/pnas.1711842115. PMC 6016768. PMID 29784790.
- ^ a b Suttle CA (2005). "Viruses in the sea". Nature. 437 (7057): 356–361. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. ISSN 0028-0836. PMID 16163346. S2CID 4370363.
- ^ "Harmful Algal Blooms: Red Tide: Home|CDC HSB". www.cdc.gov. Retrieved 19 December 2014.
- ^ a b c Suttle CA (October 2007). "Marine viruses — major players in the global ecosystem". Nature Reviews Microbiology. 5 (10): 801–812. doi:10.1038/nrmicro1750. ISSN 1740-1526. PMID 17853907. S2CID 4658457.
- ^ Breitbart M, Thompson L, Suttle C, Sullivan M (1 June 2007). "Exploring the Vast Diversity of Marine Viruses" (PDF). Oceanography. 20 (2): 135–139. doi:10.5670/oceanog.2007.58.
- ^ Hurwitz BL, U'Ren JM (June 2016). "Viral metabolic reprogramming in marine ecosystems". Current Opinion in Microbiology. 31: 161–168. doi:10.1016/j.mib.2016.04.002. PMID 27088500.
- ^ Hurwitz BL, Hallam SJ, Sullivan MB (November 2013). "Metabolic reprogramming by viruses in the sunlit and dark ocean". Genome Biology. 14 (11): R123. doi:10.1186/gb-2013-14-11-r123. PMC 4053976. PMID 24200126.
- ^ Anantharaman K, Duhaime MB, Breier JA, Wendt KA, Toner BM, Dick GJ (May 2014). "Sulfur oxidation genes in diverse deep-sea viruses". Science. 344 (6185): 757–60. Bibcode:2014Sci...344..757A. doi:10.1126/science.1252229. hdl:1912/6700. PMID 24789974. S2CID 692770.
- ^ Roux S, Hawley AK, Torres Beltran M, Scofield M, Schwientek P, Stepanauskas R, Woyke T, Hallam SJ, Sullivan MB (August 2014). "Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta-genomics". eLife. 3: e03125. doi:10.7554/elife.03125. PMC 4164917. PMID 25171894.
- ^ Forterre P, Philippe H (1999). "The last universal common ancestor (LUCA), simple or complex?". The Biological Bulletin. 196 (3): 373–5, discussion 375–7. doi:10.2307/1542973. JSTOR 1542973. PMID 11536914.
- ^ Culley A, Lang A, Suttle C (2006). "Metagenomic analysis of coastal RNA virus communities". Science. 312 (5781): 1795–1798. Bibcode:2006Sci...312.1795C. doi:10.1126/science.1127404. PMID 16794078. S2CID 20194876.
- ^ a b Rahlff J (2019). "The Virioneuston: A Review on Viral–Bacterial Associations at Air–Water Interfaces". Viruses. 11 (2): 191. doi:10.3390/v11020191. PMC 6410083. PMID 30813345..
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Hutchinson GE (1961). "The Paradox of the Plankton". The American Naturalist. 95 (882): 137–145. doi:10.1086/282171. JSTOR 2458386. S2CID 86353285.
- ^ Danovaro R, Dell'Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, Weinbauer M (August 2008). "Major viral impact on the functioning of benthic deep-sea ecosystems". Nature. 454 (7208): 1084–7. Bibcode:2008Natur.454.1084D. doi:10.1038/nature07268. PMID 18756250. S2CID 4331430.
- ^ Anderson RE, Brazelton WJ, Baross JA (2011). "Is the genetic landscape of the deep subsurface biosphere affected by viruses?". Frontiers in Microbiology. 2: 219. Bibcode:2011FrMic...2..219A. doi:10.3389/fmicb.2011.00219. ISSN 1664-302X. PMC 3211056. PMID 22084639.
- ^ Anderson RE, Sogin ML, Baross JA (3 October 2014). "Evolutionary Strategies of Viruses, Bacteria and Archaea in Hydrothermal Vent Ecosystems Revealed through Metagenomics". PLOS ONE. 9 (10): e109696. Bibcode:2014PLoSO...9j9696A. doi:10.1371/journal.pone.0109696. ISSN 1932-6203. PMC 4184897. PMID 25279954.
- ^ Ortmann AC, Suttle CA (August 2005). "High abundances of viruses in a deep-sea hydrothermal vent system indicates viral mediated microbial mortality". Deep Sea Research Part I: Oceanographic Research Papers. 52 (8): 1515–1527. Bibcode:2005DSRI...52.1515O. doi:10.1016/j.dsr.2005.04.002. ISSN 0967-0637.
- ^ Breitbart M (15 January 2012). "Marine Viruses: Truth or Dare". Annual Review of Marine Science. 4 (1): 425–448. Bibcode:2012ARMS....4..425B. doi:10.1146/annurev-marine-120709-142805. ISSN 1941-1405. PMID 22457982.
- ^ Goldenfeld N, Woese C (January 2007). "Biology's next revolution". Nature. 445 (7126): 369. arXiv:q-bio/0702015. Bibcode:2007Natur.445..369G. doi:10.1038/445369a. ISSN 0028-0836. PMID 17251963. S2CID 10737747.
- ^ a b Yau S, Seth-Pasricha M (2019). "Viruses of Polar Aquatic Environments". Viruses. 11 (2): 189. doi:10.3390/v11020189. PMC 6410135. PMID 30813316.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Anesio AM, Bellas CM (2011). "Are low temperature habitats hot spots of microbial evolution driven by viruses?". Trends in Microbiology. 19 (2): 52–57. doi:10.1016/j.tim.2010.11.002. PMID 21130655.
- ^ Laybourn-Parry J (2009). "No Place Too Cold". Science. 324 (5934): 1521–1522. Bibcode:2009Sci...324.1521L. doi:10.1126/science.1173645. PMID 19541982. S2CID 33598792.
- ^ López-Bueno A, Tamames J, Velázquez D, Moya A, Quesada A, Alcamí A (2009). "High Diversity of the Viral Community from an Antarctic Lake". Science. 326 (5954): 858–861. Bibcode:2009Sci...326..858L. doi:10.1126/science.1179287. PMID 19892985. S2CID 32607904.
- ^ Säwström C, Lisle J, Anesio AM, Priscu JC, Laybourn-Parry J (2008). "Bacteriophage in polar inland waters". Extremophiles. 12 (2): 167–175. Bibcode:2008Extmo..12..167S. doi:10.1007/s00792-007-0134-6. PMID 18188502. S2CID 2927907.
- ^ Leggett HC, Buckling A, Long GH, Boots M (October 2013). "Generalism and the evolution of parasite virulence". Trends in Ecology & Evolution. 28 (10): 592–6. Bibcode:2013TEcoE..28..592L. doi:10.1016/j.tree.2013.07.002. PMID 23968968.
- ^ Flores CO, Valverde S, Weitz JS (March 2013). "Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages". The ISME Journal. 7 (3): 520–32. Bibcode:2013ISMEJ...7..520F. doi:10.1038/ismej.2012.135. PMC 3578562. PMID 23178671.
- ^ Krupovic M, Bamford DH (July 2007). "Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria". BMC Genomics. 8: 236. doi:10.1186/1471-2164-8-236. PMC 1950889. PMID 17634101.
- ^ Xue H, Xu Y, Boucher Y, Polz MF (January 2012). "High frequency of a novel filamentous phage, VCY φ, within an environmental Vibrio cholerae population". Applied and Environmental Microbiology. 78 (1): 28–33. Bibcode:2012ApEnM..78...28X. doi:10.1128/AEM.06297-11. PMC 3255608. PMID 22020507.
- ^ a b Roux S, Krupovic M, Poulet A, Debroas D, Enault F (2012). "Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads". PLOS ONE. 7 (7): e40418. Bibcode:2012PLoSO...740418R. doi:10.1371/journal.pone.0040418. PMC 3394797. PMID 22808158.
- ^ Hurwitz BL, Sullivan MB (2013). "The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology". PLOS ONE. 8 (2): e57355. Bibcode:2013PLoSO...857355H. doi:10.1371/journal.pone.0057355. PMC 3585363. PMID 23468974.
- ^ Roux S, Enault F, Hurwitz BL, Sullivan MB (28 May 2015). "VirSorter: mining viral signal from microbial genomic data". PeerJ. 3: e985. doi:10.7717/peerj.985. PMC 4451026. PMID 26038737.
- ^ Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun F (July 2017). "VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data". Microbiome. 5 (1): 69. doi:10.1186/s40168-017-0283-5. PMC 5501583. PMID 28683828.
- ^ Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC (August 2016). "Uncovering Earth's virome". Nature. 536 (7617): 425–30. Bibcode:2016Natur.536..425P. doi:10.1038/nature19094. PMID 27533034. S2CID 4466854.
- ^ Coutinho FH, Silveira CB, Gregoracci GB, Thompson CC, Edwards RA, Brussaard CP, Dutilh BE, Thompson FL (July 2017). "Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans". Nature Communications. 8: 15955. Bibcode:2017NatCo...815955C. doi:10.1038/ncomms15955. PMC 5504273. PMID 28677677.






![Cryo-electron micrograph of the CroV giant virus [71] scale bar=0.2 μm](/uploads/wikipedia/commons/thumb/6/6e/CroV_TEM_%28cropped%29.jpg/270px-CroV_TEM_%28cropped%29.jpg?auto=webp)