Enediyne
Enediynes are organic compounds containing two triple bonds and one double bond.
Enediynes are most notable for their limited use as antitumor antibiotics (known as enediyne anticancer antibiotics).[1] They are efficient at inducing apoptosis in cells, but cannot differentiate cancerous cells from healthy cells. Consequently, research is being conducted to increase the specificity of enediyne toxicity.
Structure and reactivity
[edit]A nine- or ten-membered ring containing a double bond between two triple bonds is termed the warhead of the enediyne. In this state, the warhead is inactive. Enediynes are triggered into a chemically active state via Bergman or Myers-Saito cyclization. The triggering mechanism can be attributed to an intramolecular nucleophilic attack initiated by one of the variable regions of the molecule. Triggering can also occur via attack by an external nucleophile.[citation needed]
Bergman cyclization restructures the enediyne ring into two smaller rings. One electron from each of the enediyne triple bonds is pushed to the adjacent single bonds, generating two new double bonds. Meanwhile, another pair of electrons (one from each alkyne) is used form a new covalent bond. The resulting formation is a 1,4-benzenoid diradical fused to a ring composed of the leftover atoms from the original enediyne.

Some enediynes have an epoxide group attached to their ring, making Bergman cyclization unfavorable due to steric hindrance. For Bergman cyclization to occur, the epoxide must be removed.
Myers-Saito cyclization is another triggering mechanism by which an enediyne warhead becomes a diradical. This mechanism requires the alkene of the enediyne to be part of a diene with a double bond in a variable group. A nucleophile will attack the double bond in the variable region, causing a chain reaction of electron pushing. Ultimately, one of the triple bonds of the enediyne is converted to a cumulene.[2] The cumulene and the remaining alkyne donate one electron each to form a new covalent bond.

The diradicals generated by Bergman and Myers-Saito cyclization are highly reactive.[citation needed]
Mechanism of action
[edit]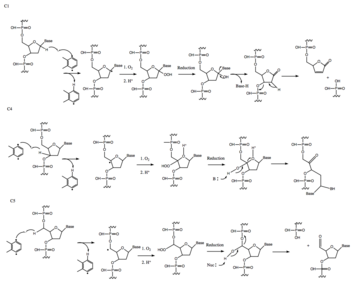
The cyclization of the enediyne functional group creates a transient reactive 1,4-benzenoid diradical that acts as a nucleophile and attacks electrophiles in order to achieve a more stable form. In biological systems, once the diradical is positioned in the minor groove of double-stranded DNA, it abstracts two hydrogen atoms from the sugars opposite strands at either the C1, C4, or C5 positions.[3] The DNA radicals that form can then cause interstrand crosslinks or react with O2, leading to double- or single-stranded DNA cleavage.[4]
Biosynthesis
[edit]Members of the enediyne family all share a unique enediyne core that is the cause of their potent cytotoxicity.[5] The enediyne cores are derived from linear, probably polyketide, precursors that consist of seven or eight head-to-tail coupled acetate units. Enediyne assembly involves a highly conserved, iterative type I polyketide synthase (PKS) pathway[6] Sequencing of enediyne gene clusters has confirmed the polyketide origin of the enediyne core, and elucidated the biosynthetic pathways and mechanisms of enediynes.[7]
In 2024, Shen and coworkers have further elucidated the biosynthesis and reported evidence for a diiodinated trienetetrayne ((13Z)-2,13-diiodopentadeca-1,7,13-triene-3,5,9,11-tetrayne) derived from pentadeca-1,3,5,7,9,11,13-heptaene as a common biosynthetic intermediate for the three known families of enediyne.[8]
Differences in the biosynthetic pathways of enediynes are due to the different origins of the -yne carbons as well as differences in isotope incorporation patterns. More differentiation comes from the attachment of various functional groups at different positions to the enediyne warheads during their maturation stage. These moieties can be either aromatic or sugars and define sequence specificity of DNA binding as well as the physical properties of the enediyne chromophores.[5]
Due to the cytotoxicity of the enediyne chromophores, their biosynthesis is tightly regulated, although the regulatory mechanisms are still largely unclear. Organisms that produce enediynes have been shown to protect themselves with a self-resistance mechanism that uses a self-sacrificing protein. Notably, some microbes use CalC to sequester calicheamicin so that the reactive diradical abstracts hydrogens from a glycine inside of the protein instead of from DNA.[5]
Classes
[edit]There are fourteen naturally occurring enediynes.[6] The other existing classes of enediynes have been synthesized in the lab.
Enediynes have been split into two sub-families: those with nine members in the core enediyne ring and those with ten-membered rings.[citation needed]
Nine-membered rings (chromoproteins)
[edit]The nine-membered enediynes are also referred to as chromoproteins because they have an attached protein as a variable group. This protein is necessary for transport and stabilization of the enediyne group.[9]
Neocarzinostatin
[edit]Neocarzinostatin is a natural product of Streptomyces carzinostaticus. It forms an apoprotein with a 113-amino acid polypeptide which can cleave histone protein H1.[10] Neocarzinostatin is an example of an enediyne that undergoes triggering via Myers-Saito cyclization. An analog of neocarzinostatin, SMANCS, has been approved for use in Japan as an antitumor drug for liver cancer.[11]
C-1027
[edit]Also known as lidamycin, C-1027 is one of the most potent antitumor enediynes. C-1027 was first isolated from Streptomyces globisporus in a soil sample taken from the Qian-Jiang District of China. Unlike most enediynes, C-1027 does not undergo a triggering process to become an activated 1,4-benzenoid diradical.[12] C-1027 has demonstrated potential efficacy against hypoxic tumors.[4]
Ten-membered rings
[edit]Calicheamicins
[edit]The calicheamicins are a sub-family of enediynes that were isolated from Micromonospora echinospora calichensis.[13] All calicheamicin family members demonstrate potent antimicrobial activity against Gram-positive and Gram-negative organisms.[13] Calicheamicin γ1 exhibited significant antitumor activity against leukemia and melanoma cells in vivo.[13] The calicheamicins are notably similar in structure to the esperamicins.
Esperamicins
[edit]The esperamicins are a sub-family of enediynes that are considered among the most potent antitumor antibiotics discovered.[14] First isolated in Actinomadura verrucosospora, members of the esperamicin family include esperamicin A1, A1b, A2, A3, A4, B1, B2, and X. Esperamicin X is an inactive esperamicin naturally produced by A. verrucosospora.[14] Compounds with thiol groups induce triggering among the esperamicins.[15]
Dynemicins
[edit]The dynemicins are a sub-family of enediynes whose members are organic compounds generated in Micromonospora chersina.[9] Dynemicin A was the first member of this sub-family to be discovered. It was isolated from M. chersina in a soil sample taken from the state of Gujarat in India.[16] Dynemicins are violet in color because they contain anthraquinone as a variable group attached to the enediyne core.[9] Dynemycins have demonstrated strong antitumor activity against leukemia and melanoma cells.[17]
Golfomycin A
[edit]Golfomycin A is a synthetic enediyne molecule designed in an attempt to create a more easily manufactured antitumor antibiotic.[18] DNA strand-scission induced by golfomycin A is pH dependent.[18] Preliminary in vitro studies have demonstrated that golfomycin A can reduce carcinomas in bladder cells.[18]
See also
[edit]References
[edit]- ^ Nicolaou KC, Smith AL, Yue EW (July 1993). "Chemistry and biology of natural and designed enediynes". Proceedings of the National Academy of Sciences of the United States of America. 90 (13): 5881–8. Bibcode:1993PNAS...90.5881N. doi:10.1073/pnas.90.13.5881. PMC 46830. PMID 8327459.
- ^ "Bergman Cyclization". www.organic-chemistry.org. Retrieved 2018-05-05.
- ^ Smith AL, Nicolaou KC (May 1996). "The enediyne antibiotics". Journal of Medicinal Chemistry. 39 (11): 2103–17. doi:10.1021/jm9600398. PMID 8667354.
- ^ a b Chen Y, Yin M, Horsman GP, Shen B (March 2011). "Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus". Journal of Natural Products. 74 (3): 420–4. doi:10.1021/np100825y. PMC 3064734. PMID 21250756.
- ^ a b c Liang ZX (April 2010). "Complexity and simplicity in the biosynthesis of enediyne natural products". Natural Product Reports. 27 (4): 499–528. doi:10.1039/b908165h. PMID 20336235.
- ^ a b Shen B, Hindra, Yan X, Huang T, Ge H, Yang D, Teng Q, Rudolf JD, Lohman JR (January 2015). "Enediynes: Exploration of microbial genomics to discover new anticancer drug leads". Bioorganic & Medicinal Chemistry Letters. 25 (1): 9–15. doi:10.1016/j.bmcl.2014.11.019. PMC 4480864. PMID 25434000.
- ^ Van Lanen SG, Shen B (2008). "Biosynthesis of enediyne antitumor antibiotics". Current Topics in Medicinal Chemistry. 8 (6): 448–59. doi:10.2174/156802608783955656. PMC 3108100. PMID 18397168.
- ^ Gui, Chun; Kalkreuter, Edward; Lauterbach, Lukas; Yang, Dong; Shen, Ben (2024). "Enediyne natural product biosynthesis unified by a diiodotetrayne intermediate". Nature Chemical Biology. 20 (9): 1210–1219. doi:10.1038/s41589-024-01636-y. ISSN 1552-4469.
- ^ a b c Gao Q, Thorson JS (May 2008). "The biosynthetic genes encoding for the production of the dynemicin enediyne core in Micromonospora chersina ATCC53710". FEMS Microbiology Letters. 282 (1): 105–14. doi:10.1111/j.1574-6968.2008.01112.x. PMC 5591436. PMID 18328078.
- ^ Heyd B, Lerat G, Adjadj E, Minard P, Desmadril M (April 2000). "Reinvestigation of the proteolytic activity of neocarzinostatin". Journal of Bacteriology. 182 (7): 1812–8. doi:10.1128/jb.182.7.1812-1818.2000. PMC 101862. PMID 10714984.
- ^ Maeda H (March 2001). "SMANCS and polymer-conjugated macromolecular drugs: advantages in cancer chemotherapy". Advanced Drug Delivery Reviews. 46 (1–3): 169–85. doi:10.1016/s0169-409x(00)00134-4. PMID 11259839.
- ^ Xu YJ, Zhen YS, Goldberg IH (May 1994). "C1027 chromophore, a potent new enediyne antitumor antibiotic, induces sequence-specific double-strand DNA cleavage". Biochemistry. 33 (19): 5947–54. doi:10.1021/bi00185a036. PMID 8180224.
- ^ a b c Maiese WM, Lechevalier MP, Lechevalier HA, Korshalla J, Kuck N, Fantini A, Wildey MJ, Thomas J, Greenstein M (April 1989). "Calicheamicins, a novel family of antitumor antibiotics: taxonomy, fermentation and biological properties". The Journal of Antibiotics. 42 (4): 558–63. doi:10.7164/antibiotics.42.558. PMID 2722671.
- ^ a b Golik J, Clardy J, Dubay G, Groenewold G, Kawaguchi H, Konishi M, Krishnan B, Ohkuma H, Saitoh K (May 1987). "Esperamicins, a novel class of potent antitumor antibiotics. 2. Structure of esperamicin X". Journal of the American Chemical Society. 109 (11): 3461–3462. doi:10.1021/ja00245a048. ISSN 0002-7863.
- ^ Sugiura Y, Uesawa Y, Takahashi Y, Kuwahara J, Golik J, Doyle TW (October 1989). "Nucleotide-specific cleavage and minor-groove interaction of DNA with esperamicin antitumor antibiotics". Proceedings of the National Academy of Sciences of the United States of America. 86 (20): 7672–6. Bibcode:1989PNAS...86.7672S. doi:10.1073/pnas.86.20.7672. PMC 298132. PMID 2813351.
- ^ Konishi M, Ohkuma H, Matsumoto K, Tsuno T, Kamei H, Miyaki T, Oki T, Kawaguchi H, VanDuyne GD, Clardy J (September 1989). "Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit". The Journal of Antibiotics. 42 (9): 1449–52. doi:10.7164/antibiotics.42.1449. PMID 2793600.
- ^ Unno R, Michishita H, Inagaki H, Suzuki Y, Baba Y, Jomori T, Nishikawa T, Isobe M (May 1997). "Synthesis and antitumor activity of water-soluble enediyne compounds related to dynemicin A". Bioorganic & Medicinal Chemistry. 5 (5): 987–99. doi:10.1016/s0968-0896(97)00037-0. PMID 9208107.
- ^ a b c Nicolaou KC, Skokotas G, Furuya S, Suemune H, Nicolaou DC (September 1990). "Golfomycin A, a Novel Designed Molecule with DNA-Cleaving Properties and Antitumor Activity". Angewandte Chemie International Edition in English. 29 (9): 1064–1067. doi:10.1002/anie.199010641. ISSN 0570-0833.


