Electrophysiology
Electrophysiology (from Greek ἥλεκτ, ēlektron, "amber" [see the etymology of "electron"]; φύσις, physis, "nature, origin"; and -λογία, -logia) is the branch of physiology that studies the electrical properties of biological cells and tissues. It involves measurements of voltage changes or electric current or manipulations on a wide variety of scales from single ion channel proteins to whole organs like the heart. In neuroscience, it includes measurements of the electrical activity of neurons, and, in particular, action potential activity. Recordings of large-scale electric signals from the nervous system, such as electroencephalography, may also be referred to as electrophysiological recordings.[1] They are useful for electrodiagnosis and monitoring.

Definition and scope
[edit]Classical electrophysiological techniques
[edit]Principle and mechanisms
[edit]Electrophysiology[2] is the branch of physiology that pertains broadly to the flow of ions (ion current) in biological tissues and, in particular, to the electrical recording techniques that enable the measurement of this flow. Classical electrophysiology techniques involve placing electrodes into various preparations of biological tissue. The principal types of electrodes are:
- Simple solid conductors, such as discs and needles (singles or arrays, often insulated except for the tip),
- Tracings on printed circuit boards or flexible polymers, also insulated except for the tip, and
- Hollow, often elongated or 'pulled', tubes filled with an electrolyte, such as glass pipettes filled with potassium chloride solution or another electrolyte solution.
The principal preparations include:
- living organisms (example in insects),
- excised tissue (acute or cultured),
- dissociated cells from excised tissue (acute or cultured),
- artificially grown cells or tissues, or
- hybrids of the above.
Neuronal electrophysiology is the study of electrical properties of biological cells and tissues within the nervous system. With neuronal electrophysiology doctors and specialists can determine how neuronal disorders happen, by looking at the individual's brain activity. Activity such as which portions of the brain light up during any situations encountered. If an electrode is small enough (micrometers) in diameter, then the electrophysiologist may choose to insert the tip into a single cell. Such a configuration allows direct observation and intracellular recording of the intracellular electrical activity of a single cell. However, this invasive setup reduces the life of the cell and causes a leak of substances across the cell membrane.
Intracellular activity may also be observed using a specially formed (hollow) glass pipette containing an electrolyte. In this technique, the microscopic pipette tip is pressed against the cell membrane, to which it tightly adheres by an interaction between glass and lipids of the cell membrane. The electrolyte within the pipette may be brought into fluid continuity with the cytoplasm by delivering a pulse of negative pressure to the pipette in order to rupture the small patch of membrane encircled by the pipette rim (whole-cell recording). Alternatively, ionic continuity may be established by "perforating" the patch by allowing exogenous pore-forming agents within the electrolyte to insert themselves into the membrane patch (perforated patch recording). Finally, the patch may be left intact (patch recording).
The electrophysiologist may choose not to insert the tip into a single cell. Instead, the electrode tip may be left in continuity with the extracellular space. If the tip is small enough, such a configuration may allow indirect observation and recording of action potentials from a single cell, termed single-unit recording. Depending on the preparation and precise placement, an extracellular configuration may pick up the activity of several nearby cells simultaneously, termed multi-unit recording.
As electrode size increases, the resolving power decreases. Larger electrodes are sensitive only to the net activity of many cells, termed local field potentials. Still larger electrodes, such as uninsulated needles and surface electrodes used by clinical and surgical neurophysiologists, are sensitive only to certain types of synchronous activity within populations of cells numbering in the millions.
Other classical electrophysiological techniques include single channel recording and amperometry.
Electrographic modalities by body part
[edit]Electrophysiological recording in general is sometimes called electrography (from electro- + -graphy, "electrical recording"), with the record thus produced being an electrogram. However, the word electrography has other senses (including electrophotography), and the specific types of electrophysiological recording are usually called by specific names, constructed on the pattern of electro- + [body part combining form] + -graphy (abbreviation ExG). Relatedly, the word electrogram (not being needed for those other senses) often carries the specific meaning of intracardiac electrogram, which is like an electrocardiogram but with some invasive leads (inside the heart) rather than only noninvasive leads (on the skin). Electrophysiological recording for clinical diagnostic purposes is included within the category of electrodiagnostic testing. The various "ExG" modes are as follows:
| Modality | Abbreviation | Body part | Prevalence in clinical use |
|---|---|---|---|
| electrocardiography | ECG or EKG | heart (specifically, the cardiac muscle), with cutaneous electrodes (noninvasive) | 1—very common |
| electroatriography | EAG | atrial cardiac muscle | 3—uncommon |
| electroventriculography | EVG | ventricular cardiac muscle | 3—uncommon |
| intracardiac electrogram | EGM | heart (specifically, the cardiac muscle), with intracardiac electrodes (invasive) | 2—somewhat common |
| electroencephalography | EEG | brain (usually the cerebral cortex), with extracranial electrodes | 2—somewhat common |
| electrocorticography | ECoG or iEEG | brain (specifically the cerebral cortex), with intracranial electrodes | 2—somewhat common |
| electromyography | EMG | muscles throughout the body (usually skeletal, occasionally smooth) | 1—very common |
| electrooculography | EOG | eye—entire globe | 2—somewhat common |
| electroretinography | ERG | eye—retina specifically | 2—somewhat common |
| electronystagmography | ENG | eye—via the corneoretinal potential | 2—somewhat common |
| electroolfactography | EOG | olfactory epithelium in mammals | 3—uncommon |
| electroantennography | EAG | olfactory receptors in arthropod antennae | 4—not applicable clinically |
| electrocochleography | ECOG or ECochG | cochlea | 2—somewhat common |
| electrogastrography | EGG | stomach smooth muscle | 2—somewhat common |
| electrogastroenterography | EGEG | stomach and bowel smooth muscle | 2—somewhat common |
| electroglottography | EGG | glottis | 3—uncommon |
| electropalatography | EPG | palatal contact of tongue | 3—uncommon |
| electroarteriography | EAG | arterial flow via streaming potential detected through skin[3] | 3—uncommon |
| electroblepharography | EBG | eyelid muscle | 3—uncommon |
| electrodermography | EDG | skin | 3—uncommon |
| electropancreatography | EPG | pancreas | 3—uncommon |
| electrohysterography | EHG | uterus | 3—uncommon |
| electroneuronography | ENeG or ENoG | nerves | 3—uncommon |
| electropneumography | EPG | lungs (chest movements) | 3—uncommon |
| electrospinography | ESG | spinal cord | 3—uncommon |
| electrovomerography | EVG | vomeronasal organ | 3—uncommon |
Optical electrophysiological techniques
[edit]Optical electrophysiological techniques were created by scientists and engineers to overcome one of the main limitations of classical techniques. Classical techniques allow observation of electrical activity at approximately a single point within a volume of tissue. Classical techniques singularize a distributed phenomenon. Interest in the spatial distribution of bioelectric activity prompted development of molecules capable of emitting light in response to their electrical or chemical environment. Examples are voltage sensitive dyes and fluorescing proteins. After introducing one or more such compounds into tissue via perfusion, injection or gene expression, the 1 or 2-dimensional distribution of electrical activity may be observed and recorded.[citation needed]
Intracellular recording
[edit]Intracellular recording involves measuring voltage and/or current across the membrane of a cell. To make an intracellular recording, the tip of a fine (sharp) microelectrode must be inserted inside the cell, so that the membrane potential can be measured. Typically, the resting membrane potential of a healthy cell will be -60 to -80 mV, and during an action potential the membrane potential might reach +40 mV. In 1963, Alan Lloyd Hodgkin and Andrew Fielding Huxley won the Nobel Prize in Physiology or Medicine for their contribution to understanding the mechanisms underlying the generation of action potentials in neurons. Their experiments involved intracellular recordings from the giant axon of Atlantic squid (Loligo pealei), and were among the first applications of the "voltage clamp" technique.[4]
Today, most microelectrodes used for intracellular recording are glass micropipettes, with a tip diameter of < 1 micrometre, and a resistance of several megohms. The micropipettes are filled with a solution that has a similar ionic composition to the intracellular fluid of the cell. A chlorided silver wire inserted into the pipette connects the electrolyte electrically to the amplifier and signal processing circuit. The voltage measured by the electrode is compared to the voltage of a reference electrode, usually a silver chloride-coated silver wire in contact with the extracellular fluid around the cell. In general, the smaller the electrode tip, the higher its electrical resistance. So an electrode is a compromise between size (small enough to penetrate a single cell with minimum damage to the cell) and resistance (low enough so that small neuronal signals can be discerned from thermal noise in the electrode tip).
Maintaining healthy brain slices is pivotal for successful electrophysiological recordings. The preparation of these slices is commonly achieved with tools such as the Compresstome vibratome, ensuring optimal conditions for accurate and reliable recordings.[5] Nevertheless, even with the highest standards of tissue handling, slice preparation induces rapid and robust phenotype changes of the brain's major immune cells, microglia, which must be taken into consideration when using this model. [6]
Voltage clamp
[edit]
The voltage clamp technique allows an experimenter to "clamp" the cell potential at a chosen value. This makes it possible to measure how much ionic current crosses a cell's membrane at any given voltage. This is important because many of the ion channels in the membrane of a neuron are voltage-gated ion channels, which open only when the membrane voltage is within a certain range. Voltage clamp measurements of current are made possible by the near-simultaneous digital subtraction of transient capacitive currents that pass as the recording electrode and cell membrane are charged to alter the cell's potential.[citation needed]
Current clamp
[edit]The current clamp technique records the membrane potential by injecting current into a cell through the recording electrode. Unlike in the voltage clamp mode, where the membrane potential is held at a level determined by the experimenter, in "current clamp" mode the membrane potential is free to vary, and the amplifier records whatever voltage the cell generates on its own or as a result of stimulation. This technique is used to study how a cell responds when electric current enters a cell; this is important for instance for understanding how neurons respond to neurotransmitters that act by opening membrane ion channels.[citation needed]
Most current-clamp amplifiers provide little or no amplification of the voltage changes recorded from the cell. The "amplifier" is actually an electrometer, sometimes referred to as a "unity gain amplifier"; its main purpose is to reduce the electrical load on the small signals (in the mV range) produced by cells so that they can be accurately recorded by low-impedance electronics. The amplifier increases the current behind the signal while decreasing the resistance over which that current passes. Consider this example based on Ohm's law: A voltage of 10 mV is generated by passing 10 nanoamperes of current across 1 MΩ of resistance. The electrometer changes this "high impedance signal" to a "low impedance signal" by using a voltage follower circuit. A voltage follower reads the voltage on the input (caused by a small current through a big resistor). It then instructs a parallel circuit that has a large current source behind it (the electrical mains) and adjusts the resistance of that parallel circuit to give the same output voltage, but across a lower resistance.
Patch-clamp recording
[edit]
This technique was developed by Erwin Neher and Bert Sakmann who received the Nobel Prize in 1991.[7] Conventional intracellular recording involves impaling a cell with a fine electrode; patch-clamp recording takes a different approach. A patch-clamp microelectrode is a micropipette with a relatively large tip diameter. The microelectrode is placed next to a cell, and gentle suction is applied through the microelectrode to draw a piece of the cell membrane (the 'patch') into the microelectrode tip; the glass tip forms a high resistance 'seal' with the cell membrane. This configuration is the "cell-attached" mode, and it can be used for studying the activity of the ion channels that are present in the patch of membrane. If more suction is now applied, the small patch of membrane in the electrode tip can be displaced, leaving the electrode sealed to the rest of the cell. This "whole-cell" mode allows very stable intracellular recording. A disadvantage (compared to conventional intracellular recording with sharp electrodes) is that the intracellular fluid of the cell mixes with the solution inside the recording electrode, and so some important components of the intracellular fluid can be diluted. A variant of this technique, the "perforated patch" technique, tries to minimize these problems. Instead of applying suction to displace the membrane patch from the electrode tip, it is also possible to make small holes on the patch with pore-forming agents so that large molecules such as proteins can stay inside the cell and ions can pass through the holes freely. Also the patch of membrane can be pulled away from the rest of the cell. This approach enables the membrane properties of the patch to be analyzed pharmacologically. Patch-clamp may also be combined with RNA sequencing in a technique known as patch-seq by extracting the cellular contents following recording in order to characterize the electrophysiological properties relationship to gene expression and cell-type.
Sharp electrode recording
[edit]In situations where one wants to record the potential inside the cell membrane with minimal effect on the ionic constitution of the intracellular fluid a sharp electrode can be used. These micropipettes (electrodes) are again like those for patch clamp pulled from glass capillaries, but the pore is much smaller so that there is very little ion exchange between the intracellular fluid and the electrolyte in the pipette. The electrical resistance of the micropipette electrode is reduced by filling with 2-4M KCl, rather than a salt concentration which mimics the intracellular ionic concentrations as used in patch clamping.[8] Often the tip of the electrode is filled with various kinds of dyes like Lucifer yellow to fill the cells recorded from, for later confirmation of their morphology under a microscope. The dyes are injected by applying a positive or negative, DC or pulsed voltage to the electrodes depending on the polarity of the dye.
Extracellular recording
[edit]Single-unit recording
[edit]An electrode introduced into the brain of a living animal will detect electrical activity that is generated by the neurons adjacent to the electrode tip. If the electrode is a microelectrode, with a tip size of about 1 micrometre, the electrode will usually detect the activity of at most one neuron. Recording in this way is in general called "single-unit" recording. The action potentials recorded are very much like the action potentials that are recorded intracellularly, but the signals are very much smaller (typically about 1 mV). Most recordings of the activity of single neurons in anesthetized and conscious animals are made in this way. Recordings of single neurons in living animals have provided important insights into how the brain processes information. For example, David Hubel and Torsten Wiesel recorded the activity of single neurons in the primary visual cortex of the anesthetized cat, and showed how single neurons in this area respond to very specific features of a visual stimulus.[9] Hubel and Wiesel were awarded the Nobel Prize in Physiology or Medicine in 1981.[10]
To prepare the brain for such electrode insertion, delicate slicing devices like the compresstome vibratome, leica vibratome, microtome are often employed. These instruments aid in obtaining precise, thin brain sections necessary for electrode placement, enabling neuroscientists to target specific brain regions for recording.[11]
Multi-unit recording
[edit]If the electrode tip is slightly larger, then the electrode might record the activity generated by several neurons. This type of recording is often called "multi-unit recording", and is often used in conscious animals to record changes in the activity in a discrete brain area during normal activity. Recordings from one or more such electrodes that are closely spaced can be used to identify the number of cells around it as well as which of the spikes come from which cell. This process is called spike sorting and is suitable in areas where there are identified types of cells with well defined spike characteristics. If the electrode tip is bigger still, in general the activity of individual neurons cannot be distinguished but the electrode will still be able to record a field potential generated by the activity of many cells.
Field potentials
[edit]
Extracellular field potentials are local current sinks or sources that are generated by the collective activity of many cells. Usually, a field potential is generated by the simultaneous activation of many neurons by synaptic transmission. The diagram to the right shows hippocampal synaptic field potentials. At the right, the lower trace shows a negative wave that corresponds to a current sink caused by positive charges entering cells through postsynaptic glutamate receptors, while the upper trace shows a positive wave that is generated by the current that leaves the cell (at the cell body) to complete the circuit. For more information, see local field potential.
Amperometry
[edit]Amperometry uses a carbon electrode to record changes in the chemical composition of the oxidized components of a biological solution. Oxidation and reduction is accomplished by changing the voltage at the active surface of the recording electrode in a process known as "scanning". Because certain brain chemicals lose or gain electrons at characteristic voltages, individual species can be identified. Amperometry has been used for studying exocytosis in the nervous and endocrine systems. Many monoamine neurotransmitters; e.g., norepinephrine (noradrenalin), dopamine, and serotonin (5-HT) are oxidizable. The method can also be used with cells that do not secrete oxidizable neurotransmitters by "loading" them with 5-HT or dopamine.
Planar patch clamp
[edit]Planar patch clamp is a novel method developed for high throughput electrophysiology.[12] Instead of positioning a pipette on an adherent cell, cell suspension is pipetted on a chip containing a microstructured aperture. A single cell is then positioned on the hole by suction and a tight connection (Gigaseal) is formed. The planar geometry offers a variety of advantages compared to the classical experiment:
- It allows for integration of microfluidics, which enables automatic compound application for ion channel screening.
- The system is accessible for optical or scanning probe techniques.
- Perfusion of the intracellular side can be performed.
-
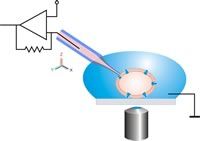
Schematic drawing of the classical patch clamp configuration. The patch pipette is moved to the cell using a micromanipulator under optical control. Relative movements between the pipette and the cell have to be avoided in order to keep the cell-pipette connection intact.
-
Scanning electron microscope image of a patch pipette.
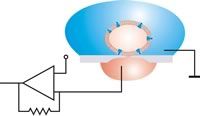
-
In planar patch configuration, the cell is positioned by suction. Relative movements between cell and aperture can then be excluded after sealing. An antivibration table is not necessary.
-
Scanning electron microscope image of a planar patch clamp chip. Both the pipette and the chip are made from borosilicate glass.
Other methods
[edit]Solid-supported membrane (SSM)-based
[edit]With this electrophysiological approach, proteoliposomes, membrane vesicles, or membrane fragments containing the channel or transporter of interest are adsorbed to a lipid monolayer painted over a functionalized electrode. This electrode consists of a glass support, a chromium layer, a gold layer, and an octadecyl mercaptane monolayer. Because the painted membrane is supported by the electrode, it is called a solid-supported membrane. Mechanical perturbations, which usually destroy a biological lipid membrane, do not influence the life-time of an SSM. The capacitive electrode (composed of the SSM and the absorbed vesicles) is so mechanically stable that solutions may be rapidly exchanged at its surface. This property allows the application of rapid substrate/ligand concentration jumps to investigate the electrogenic activity of the protein of interest, measured via capacitive coupling between the vesicles and the electrode.[13]
Bioelectric recognition assay (BERA)
[edit]The bioelectric recognition assay (BERA) is a novel method for determination of various chemical and biological molecules by measuring changes in the membrane potential of cells immobilized in a gel matrix. Apart from the increased stability of the electrode-cell interface, immobilization preserves the viability and physiological functions of the cells. BERA is used primarily in biosensor applications in order to assay analytes that can interact with the immobilized cells by changing the cell membrane potential. In this way, when a positive sample is added to the sensor, a characteristic, "signature-like" change in electrical potential occurs. BERA is the core technology behind the recently launched pan-European FOODSCAN project, about pesticide and food risk assessment in Europe.[14] BERA has been used for the detection of human viruses (hepatitis B and C viruses and herpes viruses),[15] veterinary disease agents (foot and mouth disease virus, prions, and blue tongue virus), and plant viruses (tobacco and cucumber viruses)[16] in a specific, rapid (1–2 minutes), reproducible, and cost-efficient fashion. The method has also been used for the detection of environmental toxins, such as pesticides[17][18][19] and mycotoxins[20] in food, and 2,4,6-trichloroanisole in cork and wine,[21][22] as well as the determination of very low concentrations of the superoxide anion in clinical samples.[23][24]
A BERA sensor has two parts:
- The consumable biorecognition elements
- The electronic read-out device with embedded artificial intelligence.[25]
A recent advance is the development of a technique called molecular identification through membrane engineering (MIME). This technique allows for building cells with defined specificity for virtually any molecule of interest, by embedding thousands of artificial receptors into the cell membrane.[26]
Computational electrophysiology
[edit]While not strictly constituting an experimental measurement, methods have been developed to examine the conductive properties of proteins and biomembranes in silico. These are mainly molecular dynamics simulations in which a model system like a lipid bilayer is subjected to an externally applied voltage. Studies using these setups have been able to study dynamical phenomena like electroporation of membranes[27] and ion translocation by channels.[28]
The benefit of such methods is the high level of detail of the active conduction mechanism, given by the inherently high resolution and data density that atomistic simulation affords. There are significant drawbacks, given by the uncertainty of the legitimacy of the model and the computational cost of modeling systems that are large enough and over sufficient timescales to be considered reproducing the macroscopic properties of the systems themselves. While atomistic simulations may access timescales close to, or into the microsecond domain, this is still several orders of magnitude lower than even the resolution of experimental methods such as patch-clamping.[citation needed]
Clinical electrophysiology
[edit]Clinical electrophysiology is the study of how electrophysiological principles and technologies can be applied to human health. For example, clinical cardiac electrophysiology is the study of the electrical properties which govern heart rhythm and activity. Cardiac electrophysiology can be used to observe and treat disorders such as arrhythmia (irregular heartbeat). For example, a doctor may insert a catheter containing an electrode into the heart to record the heart muscle's electrical activity.
Another example of clinical electrophysiology is clinical neurophysiology. In this medical specialty, doctors measure the electrical properties of the brain, spinal cord, and nerves. Scientists such as Duchenne de Boulogne (1806–1875) and Nathaniel A. Buchwald (1924–2006) are considered to have greatly advanced the field of neurophysiology, enabling its clinical applications.
Clinical reporting guidelines
[edit]Minimum Information (MI) standards or reporting guidelines specify the minimum amount of meta data (information) and data required to meet a specific aim or aims in a clinical study. The "Minimum Information about a Neuroscience investigation" (MINI) family of reporting guideline documents aims to provide a consistent set of guidelines in order to report an electrophysiology experiment. In practice a MINI module comprises a checklist of information that should be provided (for example about the protocols employed) when a data set is described for publication.[29]
See also
[edit]- Automated patch clamp
- Bioelectrochemistry
- Bioelectromagnetics
- Cardiac electrophysiology
- Clinical cardiac electrophysiology
- Clinical electrophysiology
- Clinical neurophysiology
- Electrophysiology study
- Hille equation
- History of bioelectricity
- Multiscale Electrophysiology Format
- Neurophysiology
- Slice preparation
- Transcutaneous electrical nerve stimulation
References
[edit]- ^ Scanziani, Massimo; Häusser, Michael (2009). "Electrophysiology in the age of light". Nature. 461 (7266): 930–39. Bibcode:2009Natur.461..930S. doi:10.1038/nature08540. PMID 19829373. S2CID 205218803.
- ^ https://www.bmseed.com/extracellular-electrophysiology
- ^ U.S. patent 4425922A
- ^ Movie featuring Alan Hodgkin recording action potentials from a squid axon https://www.youtube.com/watch?v=k48jXzFGMc8
- ^ Fontaine, R.; Hodne, K.; Weltzien, F. A. (2018). "Healthy Brain-pituitary Slices for Electrophysiological Investigations of Pituitary Cells in Teleost Fish". Journal of Visualized Experiments (138). doi:10.3791/57790. PMC 6126815. PMID 30176004.
- ^ Peter, Berki; Csaba, Cserep; Zsuzsanna, Környei (2024). "Microglia contribute to neuronal synchrony despite endogenous ATP-related phenotypic transformation in acute mouse brain slices". Nature Communications. 15 (1): 5402. Bibcode:2024NatCo..15.5402B. doi:10.1038/s41467-024-49773-1. PMC 11208608. PMID 38926390.
- ^ "The Nobel Prize in Physiology or Medicine 1991". nobelprize.org. Archived from the original on 10 October 2017. Retrieved 5 May 2018.
- ^ Halliwell J., Whitaker M., Ogden D. (1994) Using microelectrodes. Microelectrode techniques: the Plymouth Workshop handbook. ed. Ogden, D. Available online at http://plymsea.ac.uk/id/eprint/7954/
- ^ D. H. Hubel; Wiesel, TN (1 January 1962). "Receptive fields, binocular interaction and functional architecture in the cat's visual cortex". The Journal of Physiology. 160 (1): 106–54. doi:10.1113/jphysiol.1962.sp006837. PMC 1359523. PMID 14449617.
- ^ "The Nobel Prize in Physiology or Medicine 1981". nobelprize.org. Archived from the original on 23 December 2017. Retrieved 5 May 2018.
- ^ Papouin, T.; Haydon, P. G. (2018). "Obtaining Acute Brain Slices". Bio-Protocol. 8 (2): e2699. PMC 5856250. PMID 29552595.
- ^ "Automated patch clamp" (PDF). Archived (PDF) from the original on 31 March 2010. Retrieved 17 January 2010.
- ^ Schulz, Patrick; Garcia-Celma, Juan J.; Fendler, Klaus (2008). "SSM-based electrophysiology". Methods. 46 (2): 97–103. doi:10.1016/j.ymeth.2008.07.002. PMID 18675360.
- ^ Kintzios S., E. Pistola, P. Panagiotopoulos, M. Bomsel, N. Alexandropoulos, F. Bem, I. Biselis, R. Levin (2001) Bioelectric recognition assay (BERA). Biosensors and Bioelectronics 16: 325–36
- ^ Perdikaris, A.; Alexandropoulos, N; Kintzios, S. (2009) Development of a Novel, Ultra-rapid Biosensor for the Qualitative Detection of Hepatitis B Virus-associated Antigens and Anti-HBV, Based on "Membrane-engineered" Fibroblast Cells with Virus-Specific Antibodies and Antigens. Sensors 9: 2176–86.
- ^ Moschopoulou G.; Vitsa, K.; Bem, F.; Vassilakos, N.; Perdikaris, A.; Blouhos, P.; Yialouris, C.; Frossiniotis, D.; Anthopoulos, I.; Maggana, O.; Nomikou, K.; Rodeva, V.; Kostova, D.; Grozeva, S.; Michaelides, A.; Simonian, A.; Kintzios, S. (2008) Engineering of the membrane of fibroblast cells with virus-specific antibodies: a novel biosensor tool for virus detection. Biosensors Bioelectron. 24: 1033–36.
- ^ Flampouri E, Mavrikou S, Kintzios S, Miliaids G, Aplada-Sarli P (2010). Development and Validation of a Cellular Biosensor Detecting Pesticide Residues in Tomatoes. Talanta 80: 1799–804.
- ^ Mavrikou, S, Flampouri, E, Moschopoulou, G, Mangana, O, Michaelides, A, Kintzios, S (2008) Assessment of organophosphate and carbamate pesticide residues in cigarette tobacco with a novel cell biosensor. Sensors 8: 2818–32
- ^ Lokka K., Skandamis P., Kintzios S. (2013) Screening of Total Organophosphate Pesticides in Agricultural Products with a Cellular Biosensor CellBio 2: 131–37.
- ^ Larou, E., Yiakoumettis, I., Kaltsas, G., Petropoulos, A., Skandamis, P., Kintzios, S. (2012) High throughput cellular biosensor for the ultra-sensitive, ultra-rapid detection of aflatoxin M1. Food Control 29: 208–12
- ^ Varelas, V., Sanvicens N, Marco MP, Kintzios S (2010) Development of a cellular biosensor for the detection of 2, 4, 6- trichloroanisole (TCA). Talanta 84: 936–40
- ^ Apostolou T, Pascual N, Marco M-P, Moschos A, Petropoulos A, Kaltsas G, Kintzios S (2014) Extraction-less, rapid assay for the direct detection of 2,4,6-trichloroanisole (TCA) in cork samples. Talanta 125: 336–40.
- ^ Moschopoulou G., Kintzios S. (2006) Application of "membrane-engineering" to bioelectric recognition cell sensors for the detection of picomole concentrations of superoxide radical: a novel biosensor principle. Anal. Chimica Acta 573–74: 90–96.
- ^ Moschopoulou, G., Valero, T., Kintzios, S. (2012) Superoxide determination using membrane-engineered cells: An example of a novel concept for the construction of cell sensors with customized target recognition properties. Sens. Actuat.175: 88–94
- ^ Ferentinos K.P., C.P. Yialouris, P. Blouchos, G. Moschopoulou, V. Tsourou, Kintzios, S. (2013) Pesticide Residue Screening Using a Novel Artificial Neural Network Combined with a Bioelectric Cellular Biosensor. BioMed Research International. Article ID 813519.
- ^ Kokla A, Blouchos P., Livaniou E., Zikos C., Kakabakos S.E., Petrou P.S., Kintzios, S. (2013) Visualization of the membrane-engineering concept: evidence for the specific orientation of electroinserted antibodies and selective binding of target analytes. Journal of Molecular Recognition 26: 627–232.
- ^ Gurtovenko, Andrey A.; Vattulainen, Ilpo (2007). "Ion Leakage through Transient Water Pores in Protein-Free Lipid Membranes Driven by Transmembrane Ionic Charge Imbalance". Biophysical Journal. 92 (6): 1878–90. Bibcode:2007BpJ....92.1878G. doi:10.1529/biophysj.106.094797. PMC 1861780. PMID 17208976.
- ^ Kutzner, Carsten; Grubmüller, Helmut; De Groot, Bert L.; Zachariae, Ulrich (2011). "Computational Electrophysiology: The Molecular Dynamics of Ion Channel Permeation and Selectivity in Atomistic Detail". Biophysical Journal. 101 (4): 809–17. Bibcode:2011BpJ...101..809K. doi:10.1016/j.bpj.2011.06.010. PMC 3175076. PMID 21843471.
- ^ Gibson, Frank; Overton, Paul G.; Smulders, Tom V.; Schultz, Simon R.; Eglen, Stephen J.; Ingram, Colin D.; Panzeri, Stefano; Bream, Phil; Sernagor, Evelyne (2008). "Minimum Information about a Neuroscience Investigation (MINI) Electrophysiology" (PDF). Nature Precedings. hdl:10101/npre.2009.1720.2.
External links
[edit]- Book chapter on Planar Patch Clamp Archived 31 March 2010 at the Wayback Machine