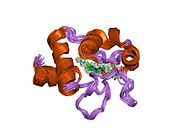
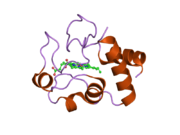
Cytochrome c

The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III (Coenzyme Q – Cyt c reductase) and IV (Cyt c oxidase). Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.[5][6]
Species distribution
[edit]Cytochrome c is a highly conserved protein across the spectrum of eukaryotic species, found in plants, animals, fungi, and many unicellular organisms. This, along with its small size (molecular weight about 12,000 daltons),[7] makes it useful in studies of cladistics.[8] Cytochrome c has been studied for the glimpse it gives into evolutionary biology.
Cytochrome c has a primary structure consisting of a chain of about 100 amino acids. Many higher-order organisms possess a chain of 104 amino acids.[9] The sequence of cytochrome c in humans is identical to that of chimpanzees (our closest relatives), but differs from that of horses.[10]
Cytochrome c has an amino acid sequence that is highly conserved in eukaryotes, varying by only a few residues. In more than thirty species tested in one study, 34 of the 104 amino acids were conserved (identical at their characteristic position).[11] For example, human cytochrome oxidase reacted with wheat cytochrome c, in vitro; which held true for all pairs of species tested.[11] In addition, the redox potential of +0.25 volts is the same in all cytochrome c molecules studied.[11]
Structure
[edit]
Cytochrome c belongs to class I of the c-type cytochrome family[13] and contains a characteristic CXXCH (cysteine-any-any-cysteine-histidine) amino acid motif that binds heme.[14] This motif is located towards the N-terminus of the peptide chain and contains a histidine as the 5th ligand of the heme iron. The 6th ligand is provided by a methionine residue found towards the C-terminus. The protein backbone is folded into five α-helices that are numbered α1-α5 from N-terminus to C-terminus. Helices α3, α4 and α5 are referred to as 50s, 60s and 70s helices, respectively, when referring to mitochondrial cytochrome c.[15]
Heme c
[edit]
While most heme proteins are attached to the prosthetic group through iron ion ligation and tertiary interactions, the heme group of cytochrome c makes thioether bonds with two cysteine side chains of the protein.[16] One of the main properties of heme c, which allows cytochrome c to have variety of functions, is its ability to have different reduction potentials in nature. This property determines the kinetics and thermodynamics of an electron transfer reaction.[17]
Dipole moment
[edit]The dipole moment has an important role in orienting proteins to the proper directions and enhancing their abilities to bind to other molecules.[18][19] The dipole moment of cytochrome c results from a cluster of negatively charged amino acid side chains at the "back" of the enzyme.[19] Despite variations in the number of bound heme groups and variations in sequence, the dipole moment of vertebrate cytochromes c is remarkably conserved. For example, vertebrate cytochromes c all have a dipole moment of approximately 320 debye while cytochromes c of plants and insects have a dipole moment of approximately 340 debye.[19]
Function
[edit]Electron transport chain
[edit]Cytochrome c is an essential component of the respiratory electron transport chain in mitochondria. The heme group of cytochrome c accepts electrons from the bc1 Complex III and transports them to Complex IV, while it transfers energy in the opposite direction. [citation needed]
Cytochrome c can also catalyze several redox reactions such as hydroxylation and aromatic oxidation, and shows peroxidase activity by oxidation of various electron donors such as 2,2-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS), 2-keto-4-thiomethyl butyric acid and 4-aminoantipyrine. [citation needed]
A bacterial cytochrome c functions as a nitrite reductase.[20]
Role in apoptosis
[edit]Cytochrome c was also discovered in 1996 by Xiaodong Wang to have an intermediate role in apoptosis, a controlled form of cell death used to kill cells in the process of development or in response to infection or DNA damage.[21]
Cytochrome c binds to cardiolipin in the inner mitochondrial membrane, thus anchoring its presence and keeping it from releasing out of the mitochondria and initiating apoptosis. While the initial attraction between cardiolipin and cytochrome c is electrostatic due to the extreme positive charge on cytochrome c, the final interaction is hydrophobic, where a hydrophobic tail from cardiolipin inserts itself into the hydrophobic portion of cytochrome c. [citation needed]
During the early phase of apoptosis, mitochondrial ROS production is stimulated, and cardiolipin is oxidized by a peroxidase function of the cardiolipin–cytochrome c complex. The hemoprotein is then detached from the mitochondrial inner membrane and can be extruded into the soluble cytoplasm through pores in the outer membrane.[22]
The sustained elevation in calcium levels precedes cyt c release from the mitochondria. The release of small amounts of cyt c leads to an interaction with the IP3 receptor (IP3R) on the endoplasmic reticulum (ER), causing ER calcium release. The overall increase in calcium triggers a massive release of cyt c, which then acts in the positive feedback loop to maintain ER calcium release through the IP3Rs.[23] This explains how the ER calcium release can reach cytotoxic levels. This release of cytochrome c in turn activates caspase 9, a cysteine protease. Caspase 9 can then go on to activate caspase 3 and caspase 7, which are responsible for destroying the cell from within. [citation needed]
Inhibition of apoptosis
[edit]One of the ways cell apoptosis is activated is by release of cytochrome c from the mitochondria into cytosol. A study has shown that cells are able to protect themselves from apoptosis by blocking the release of cytochrome c using Bcl-xL.[24] Another way that cells can control apoptosis is by phosphorylation of Tyr48, which would turn cytochrome c into an anti-apoptotic switch.[25]
As an antioxidative enzyme
[edit]
In addition to its well-known roles in the electron transport chain and cell apoptosis, according to a recent study cytochrome c can also act as an antioxidative enzyme in the mitochondria; it does so by removing superoxide (O−2) and hydrogen peroxide (H2O2) from mitochondria.[26] Therefore, not only is cytochrome c required in the mitochondria for cellular respiration, but it is also needed in the mitochondria to limit the production of O−2 and H2O2.[26]
Extramitochondrial localisation
[edit]Cytochrome c is widely believed to be localised solely in the mitochondrial intermembrane space under normal physiological conditions.[27] The release of cytochrome c from mitochondria to the cytosol, where it activates the caspase family of proteases, is believed to be the primary trigger leading to the onset of apoptosis.[28] Measuring the amount of cytochrome c leaking from mitochondria to cytosol, and out of the cell to culture medium, is a sensitive method to monitor the degree of apoptosis.[29][30] However, detailed immuno-electronmicroscopic studies with rat tissues sections employing cytochrome c specific antibodies provide compelling evidence that cytochrome c under normal cellular conditions is also present at extramitochondrial locations.[31] In pancreatic acinar cells and the anterior pituitary, strong and specific presence of cytochrome c was detected in zymogen granules and in growth hormone granules, respectively. In the pancreas, cytochrome c was also found in condensing vacuoles and in the acinar lumen. The extramitochondrial localisation of cytochrome c was shown to be specific as it was completely abolished upon adsorption of the primary antibody with purified cytochrome c.[31] Besides cytochrome c, extramitochondrial localisation has also been observed for large numbers of other proteins including those encoded by mitochondrial DNA.[32][33][34] This raises the possibility of the existence of yet-unidentified specific mechanisms for protein translocation from mitochondria to other cellular destinations.[34][35]
Applications
[edit]Superoxide detection
[edit]
Cytochrome c has been used to detect peroxide production in biological systems. As superoxide is produced, the number of oxidised cytochrome c3+ increases, and reduced cytochrome c2+ decreases.[36] However, superoxide is often produced with nitric oxide. In the presence of nitric oxide, the reduction of cytochrome c3+ is inhibited.[37] This leads to the oxidisation of cytochrome c2+ to cytochrome c3+ by peroxynitrous acid, an intermediate made through the reaction of nitric oxide and superoxide.[37] Presence of peroxynitrite or H2O2 and nitrogen dioxide NO2 in the mitochondria can be lethal since they nitrate tyrosine residues of cytochrome c, which leads to disruption of cytochrome c's function as an electron carrier in the electron transport chain.[38]
As an enzyme for Catalytic Activity
[edit]Cytochrome C has also been widely studied as an enzyme with peroxidase-like activity. Cytochrome C was conjugated to charged polymer to test its peroxidase-like activity.[39][40] Inspired from natural examples of enzyme encapsulation in protein-based cage structures (Example: Carboxysomes, Ferritin and Encapsulin), Cytochrome C was encapsulated in a 9 nm small self-assembling DNA binding protein from nutrient starved cells (Dps) protein cage using chimeric self-assembly approach. Authors observed unique catalytic activity behavior upon encapsulating enzyme inside a protein-cage, which was different from enzyme in solution. This was attributed to local microenvironment provided by Dps nanocage's interior cavity which is different than bulk.[41]
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000172115 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000063694 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: cytochrome c".
- ^ Tafani M, Karpinich NO, Hurster KA, Pastorino JG, Schneider T, Russo MA, et al. (March 2002). "Cytochrome c release upon Fas receptor activation depends on translocation of full-length bid and the induction of the mitochondrial permeability transition". The Journal of Biological Chemistry. 277 (12): 10073–82. doi:10.1074/jbc.M111350200. PMID 11790791.
- ^ "Cytochrome c – Homo sapiens (Human)". P99999. UniProt Consortium.
mass is 11,749 Daltons
- ^ Margoliash E (October 1963). "Primary structure and evolution of cytochrome c". Proceedings of the National Academy of Sciences of the United States of America. 50 (4): 672–9. Bibcode:1963PNAS...50..672M. doi:10.1073/pnas.50.4.672. PMC 221244. PMID 14077496.
- ^ "Amino acid sequences in cytochrome c proteins from different species" (PDF). Archived from the original (PDF) on 2013-12-28., adapted from Strahler AN (1999). Science and earth history: the evolution/creation controversy. Amherst, N.Y: Prometheus Books. p. 348. ISBN 978-1-57392-717-8.
- ^ Lurquin PF, Stone L, Cavalli-Sforza LL (2007). Genes, culture, and human evolution: a synthesis. Oxford: Blackwell. p. 79. ISBN 978-1-4051-5089-7.
- ^ a b c Stryer L (1975). Biochemistry (1st ed.). San Francisco: W.H. Freeman and Company. p. 362. ISBN 978-0-7167-0174-3.
- ^ McPherson A, DeLucas LJ (2015). "Microgravity protein crystallization". npj Microgravity. 1: 15010. doi:10.1038/npjmgrav.2015.10. PMC 5515504. PMID 28725714.
- ^ Ambler RP (May 1991). "Sequence variability in bacterial cytochromes c". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1058 (1): 42–7. doi:10.1016/S0005-2728(05)80266-X. PMID 1646017.
- ^ Mavridou DA, Ferguson SJ, Stevens JM (March 2013). "Cytochrome c assembly". IUBMB Life. 65 (3): 209–16. doi:10.1002/iub.1123. PMID 23341334. S2CID 32216217.
- ^ Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, et al. (2014-04-23). "Metalloproteins Containing Cytochrome, Iron–Sulfur, or Copper Redox Centers". Chemical Reviews. 114 (8): 4366–4469. doi:10.1021/cr400479b. ISSN 0009-2665. PMC 4002152. PMID 24758379.
- ^ Kang X, Carey J (November 1999). "Role of heme in structural organization of cytochrome c probed by semisynthesis". Biochemistry. 38 (48): 15944–51. doi:10.1021/bi9919089. PMID 10625461.
- ^ Zhao Y, Wang ZB, Xu JX (January 2003). "Effect of cytochrome c on the generation and elimination of O2– and H2O2 in mitochondria". The Journal of Biological Chemistry. 278 (4): 2356–60. doi:10.1074/jbc.M209681200. PMID 12435729.
- ^ Koppenol WH, Margoliash E (April 1982). "The asymmetric distribution of charges on the surface of horse cytochrome c. Functional implications". The Journal of Biological Chemistry. 257 (8): 4426–37. doi:10.1016/S0021-9258(18)34740-9. PMID 6279635.
- ^ a b c Koppenol WH, Rush JD, Mills JD, Margoliash E (July 1991). "The dipole moment of cytochrome c". Molecular Biology and Evolution. 8 (4): 545–58. doi:10.1093/oxfordjournals.molbev.a040659. PMID 1656165.
- ^ Schneider J, Kroneck PM (2014). "The Production of Ammonia by Multiheme Cytochromes C". In Kroneck PM, Torres ME (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. Vol. 14. Springer. pp. 211–236. doi:10.1007/978-94-017-9269-1_9. ISBN 978-94-017-9268-4. PMID 25416396.
- ^ Liu X, Kim CN, Yang J, Jemmerson R, Wang X (July 1996). "Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c". Cell. 86 (1): 147–57. doi:10.1016/S0092-8674(00)80085-9. PMID 8689682. S2CID 12604356.
- ^ Orrenius S, Zhivotovsky B (September 2005). "Cardiolipin oxidation sets cytochrome c free". Nature Chemical Biology. 1 (4): 188–9. doi:10.1038/nchembio0905-188. PMID 16408030. S2CID 45381495.
- ^ Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH (December 2003). "Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis". Nature Cell Biology. 5 (12): 1051–61. doi:10.1038/ncb1063. PMID 14608362. S2CID 27761335.
- ^ Kharbanda S, Pandey P, Schofield L, Israels S, Roncinske R, Yoshida K, et al. (June 1997). "Role for Bcl-xL as an inhibitor of cytosolic cytochrome C accumulation in DNA damage-induced apoptosis". Proceedings of the National Academy of Sciences of the United States of America. 94 (13): 6939–42. Bibcode:1997PNAS...94.6939K. doi:10.1073/pnas.94.13.6939. PMC 21263. PMID 9192670.
- ^ García-Heredia JM, Díaz-Quintana A, Salzano M, Orzáez M, Pérez-Payá E, Teixeira M, et al. (December 2011). "Tyrosine phosphorylation turns alkaline transition into a biologically relevant process and makes human cytochrome c behave as an anti-apoptotic switch". Journal of Biological Inorganic Chemistry. 16 (8): 1155–68. doi:10.1007/s00775-011-0804-9. PMID 21706253. S2CID 24156094.
- ^ a b Bowman SE, Bren KL (December 2008). "The chemistry and biochemistry of heme c: functional bases for covalent attachment". Natural Product Reports. 25 (6): 1118–30. doi:10.1039/b717196j. PMC 2654777. PMID 19030605.
- ^ Neupert W (1997). "Protein import into mitochondria". Annual Review of Biochemistry. 66: 863–917. doi:10.1146/annurev.biochem.66.1.863. PMID 9242927.
- ^ Kroemer G, Dallaporta B, Resche-Rigon M (1998). "The mitochondrial death/life regulator in apoptosis and necrosis". Annual Review of Physiology. 60: 619–42. doi:10.1146/annurev.physiol.60.1.619. PMID 9558479.
- ^ Loo JF, Lau PM, Ho HP, Kong SK (October 2013). "An aptamer-based bio-barcode assay with isothermal recombinase polymerase amplification for cytochrome-c detection and anti-cancer drug screening". Talanta. 115: 159–65. doi:10.1016/j.talanta.2013.04.051. PMID 24054573.
- ^ Waterhouse NJ, Trapani JA (July 2003). "A new quantitative assay for cytochrome c release in apoptotic cells". Cell Death and Differentiation. 10 (7): 853–5. doi:10.1038/sj.cdd.4401263. PMID 12815469.
- ^ a b Soltys BJ, Andrews DW, Jemmerson R, Gupta RS (2001). "Cytochrome-C localises in secretory granules in pancreas and anterior pituitary". Cell Biology International. 25 (4): 331–8. doi:10.1006/cbir.2000.0651. PMID 11319839. S2CID 2106599.
- ^ Gupta RS, Ramachandra NB, Bowes T, Singh B (2008). "Unusual Cellular Disposition of the Mitochondrial Molecular Chaperones Hsp60, Hsp70 and Hsp10". In Chadwick D, Goode J (eds.). The Biology of Extracellular Molecular Chaperones. Novartis Foundation Symposia. Vol. 291. pp. 59–68, discussion 69–73, 137–40. doi:10.1002/9780470754030.ch5. ISBN 978-0-470-75403-0. PMID 18575266.
- ^ Sadacharan SK, Singh B, Bowes T, Gupta RS (November 2005). "Localisation of mitochondrial DNA encoded cytochrome c oxidase subunits I and II in rat pancreatic zymogen granules and pituitary growth hormone granules". Histochemistry and Cell Biology. 124 (5): 409–21. doi:10.1007/s00418-005-0056-2. PMID 16133117. S2CID 24440427.
- ^ a b Soltys BJ, Gupta RS (2000). Mitochondrial proteins at unexpected cellular locations: export of proteins from mitochondria from an evolutionary perspective. International Review of Cytology. Vol. 194. pp. 133–96. doi:10.1016/s0074-7696(08)62396-7. ISBN 978-0-12-364598-2. PMID 10494626.
- ^ Soltys BJ, Gupta RS (May 1999). "Mitochondrial-matrix proteins at unexpected locations: are they exported?". Trends in Biochemical Sciences. 24 (5): 174–7. doi:10.1016/s0968-0004(99)01390-0. PMID 10322429.
- ^ McCord JM, Fridovich I (November 1969). "Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein)". The Journal of Biological Chemistry. 244 (22): 6049–55. doi:10.1016/S0021-9258(18)63504-5. PMID 5389100.
- ^ a b Thomson L, Trujillo M, Telleri R, Radi R (June 1995). "Kinetics of cytochrome c2+ oxidation by peroxynitrite: implications for superoxide measurements in nitric oxide-producing biological systems". Archives of Biochemistry and Biophysics. 319 (2): 491–7. doi:10.1006/abbi.1995.1321. PMID 7786032.
- ^ Domazou AS, Gebicka L, Didik J, Gebicki JL, van der Meijden B, Koppenol WH (April 2014). "The kinetics of the reaction of nitrogen dioxide with iron(II)- and iron(III) cytochrome c". Free Radical Biology & Medicine. 69: 172–80. doi:10.1016/j.freeradbiomed.2014.01.014. PMID 24447894.
- ^ Zhang Y, Wang Q, Hess H (March 2017). "Increasing enzyme cascade throughput by pH-engineering the microenvironment of individual enzymes". ACS Catalysis. 7 (3): 2047–2051. doi:10.1021/acscatal.7b01766.
- ^ Benson KR, Gorecki J, Nikiforov A, Tsui W, Kasi RM, Kumar CV (April 2019). "Cytochrome c-poly(acrylic acid) conjugates with improved peroxidase turnover number". Organic & Biomolecular Chemistry. 17 (16): 4043–4048. doi:10.1039/c9ob00541b. PMID 30950479.
- ^ Waghwani HK, Douglas, T (March 2021). "Cytochrome C with peroxidase-like activity encapsulated inside the small DPS protein nanocage". Journal of Materials Chemistry B. 9 (14): 3168–3179. doi:10.1039/d1tb00234a. PMID 33885621.
Further reading
[edit]- Kumarswamy R, Chandna S (February 2009). "Putative partners in Bax mediated cytochrome-c release: ANT, CypD, VDAC or none of them?". Mitochondrion. 9 (1): 1–8. doi:10.1016/j.mito.2008.10.003. PMID 18992370.
- Skulachev VP (February 1998). "Cytochrome c in the apoptotic and antioxidant cascades". FEBS Letters. 423 (3): 275–80. doi:10.1016/S0014-5793(98)00061-1. PMID 9515723. S2CID 10267410.
- Mannella CA (1998). "Conformational changes in the mitochondrial channel protein, VDAC, and their functional implications". Journal of Structural Biology. 121 (2): 207–18. doi:10.1006/jsbi.1997.3954. PMID 9615439.
- Ferri KF, Jacotot E, Blanco J, Esté JA, Kroemer G (2000). "Mitochondrial control of cell death induced by HIV-1-encoded proteins". Annals of the New York Academy of Sciences. 926 (1): 149–64. Bibcode:2000NYASA.926..149F. doi:10.1111/j.1749-6632.2000.tb05609.x. PMID 11193032. S2CID 21997163.
- Britton RS, Leicester KL, Bacon BR (October 2002). "Iron toxicity and chelation therapy". International Journal of Hematology. 76 (3): 219–28. doi:10.1007/BF02982791. PMID 12416732. S2CID 22572183.
- Haider N, Narula N, Narula J (December 2002). "Apoptosis in heart failure represents programmed cell survival, not death, of cardiomyocytes and likelihood of reverse remodeling". Journal of Cardiac Failure. 8 (6 Suppl): S512–7. doi:10.1054/jcaf.2002.130034. PMID 12555167.
- Castedo M, Perfettini JL, Andreau K, Roumier T, Piacentini M, Kroemer G (December 2003). "Mitochondrial apoptosis induced by the HIV-1 envelope". Annals of the New York Academy of Sciences. 1010 (1): 19–28. Bibcode:2003NYASA1010...19C. doi:10.1196/annals.1299.004. PMID 15033690. S2CID 37073602.
- Ng S, Smith MB, Smith HT, Millett F (November 1977). "Effect of modification of individual cytochrome c lysines on the reaction with cytochrome b5". Biochemistry. 16 (23): 4975–8. doi:10.1021/bi00642a006. PMID 199233.
- Lynch SR, Sherman D, Copeland RA (January 1992). "Cytochrome c binding affects the conformation of cytochrome a in cytochrome c oxidase". The Journal of Biological Chemistry. 267 (1): 298–302. doi:10.1016/S0021-9258(18)48493-1. PMID 1309738.
- Garber EA, Margoliash E (February 1990). "Interaction of cytochrome c with cytochrome c oxidase: an understanding of the high- to low-affinity transition". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1015 (2): 279–87. doi:10.1016/0005-2728(90)90032-Y. PMID 2153405.
- Bedetti CD (May 1985). "Immunocytochemical demonstration of cytochrome c oxidase with an immunoperoxidase method: a specific stain for mitochondria in formalin-fixed and paraffin-embedded human tissues". The Journal of Histochemistry and Cytochemistry. 33 (5): 446–52. doi:10.1177/33.5.2580882. PMID 2580882.
- Tanaka Y, Ashikari T, Shibano Y, Amachi T, Yoshizumi H, Matsubara H (June 1988). "Construction of a human cytochrome c gene and its functional expression in Saccharomyces cerevisiae". Journal of Biochemistry. 103 (6): 954–61. doi:10.1093/oxfordjournals.jbchem.a122393. PMID 2844747.
- Evans MJ, Scarpulla RC (December 1988). "The human somatic cytochrome c gene: two classes of processed pseudogenes demarcate a period of rapid molecular evolution". Proceedings of the National Academy of Sciences of the United States of America. 85 (24): 9625–9. Bibcode:1988PNAS...85.9625E. doi:10.1073/pnas.85.24.9625. PMC 282819. PMID 2849112.
- Passon PG, Hultquist DE (July 1972). "Soluble cytochrome b 5 reductase from human erythrocytes". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 275 (1): 62–73. doi:10.1016/0005-2728(72)90024-2. hdl:2027.42/34070. PMID 4403130.
- Dowe RJ, Vitello LB, Erman JE (August 1984). "Sedimentation equilibrium studies on the interaction between cytochrome c and cytochrome c peroxidase". Archives of Biochemistry and Biophysics. 232 (2): 566–73. doi:10.1016/0003-9861(84)90574-5. PMID 6087732.
- Michel B, Bosshard HR (August 1984). "Spectroscopic analysis of the interaction between cytochrome c and cytochrome c oxidase". The Journal of Biological Chemistry. 259 (16): 10085–91. doi:10.1016/S0021-9258(18)90932-4. PMID 6088481.
- Broger C, Nałecz MJ, Azzi A (October 1980). "Interaction of cytochrome c with cytochrome bc1 complex of the mitochondrial respiratory chain". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 592 (3): 519–27. doi:10.1016/0005-2728(80)90096-1. PMID 6251869.
- Smith HT, Ahmed AJ, Millett F (May 1981). "Electrostatic interaction of cytochrome c with cytochrome c1 and cytochrome oxidase". The Journal of Biological Chemistry. 256 (10): 4984–90. doi:10.1016/S0021-9258(19)69355-5. PMID 6262312.
- Geren LM, Millett F (October 1981). "Fluorescence energy transfer studies of the interaction between adrenodoxin and cytochrome c". The Journal of Biological Chemistry. 256 (20): 10485–9. doi:10.1016/S0021-9258(19)68647-3. PMID 6270113.
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA (June 1994). "The catalytic subunit of protein phosphatase 2A is carboxyl-methylated in vivo". The Journal of Biological Chemistry. 269 (23): 16311–7. doi:10.1016/S0021-9258(17)34009-7. PMID 8206937.
- Gao B, Eisenberg E, Greene L (July 1996). "Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate". The Journal of Biological Chemistry. 271 (28): 16792–7. doi:10.1074/jbc.271.28.16792. PMID 8663341.
External links
[edit]- Apoptosis & Caspase 3 – PMAP The Proteolysis Map-animation
- Cytochrome+c at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P99999 (Cytochrome c) at the PDBe-KB.