Clostridium perfringens
| Clostridium perfringens | |
|---|---|
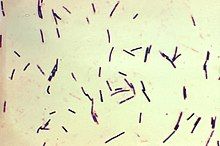
| |
| Photomicrograph of Gram-positive Clostridium perfringens bacilli | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Bacillota |
| Class: | Clostridia |
| Order: | Eubacteriales |
| Family: | Clostridiaceae |
| Genus: | Clostridium |
| Species: | C. perfringens
|
| Binomial name | |
| Clostridium perfringens Veillon & Zuber 1898
Hauduroy et al. 1937 | |
Clostridium perfringens (formerly known as C. welchii, or Bacillus welchii) is a Gram-positive, bacillus (rod-shaped), anaerobic, spore-forming pathogenic bacterium of the genus Clostridium.[1][2] C. perfringens is ever-present in nature and can be found as a normal component of decaying vegetation, marine sediment, the intestinal tract of humans and other vertebrates, insects, and soil. It has the shortest reported generation time of any organism at 6.3 minutes in thioglycolate medium.[3]
Clostridium perfringens is one of the most common causes of food poisoning in the United States, alongside norovirus, Salmonella, Campylobacter, and Staphylococcus aureus.[4] However, it can sometimes be ingested and cause no harm.[5]
Infections induced by C. perfringens are associated with tissue necrosis, bacteremia, emphysematous cholecystitis, and gas gangrene, which is also known as clostridial myonecrosis.[6] The specific name, perfringens, is derived from the Latin per (meaning "through") and frango ("burst"), referring to the disruption of tissue that occurs during gas gangrene.[7] Gas gangrene is caused by alpha toxin, or α-toxin, that embeds itself into the plasma membrane of cells and disrupts normal cellular function by altering membrane structure.[8] Research suggests that C. perfringens is capable of engaging in polymicrobial anaerobic infections.[9] It is commonly encountered in infections as a component of the normal flora. In this case, its role in disease is minor.[10]
C. perfringens toxins are a result of horizontal gene transfer of a neighboring cell's plasmids.[11] Shifts in genomic make-up are common for this species of bacterium and contribute to novel pathogenesis.[12] Major toxins are expressed differently in certain populations of C. perfringens; these populations are organized into strains based on their expressed toxins.[13] This especially impacts the food industry, as controlling this microbe is important for preventing foodborne illness.[12] Novel findings in C. perfringens hyper-motility, which was provisionally thought as non-motile, have been discovered as well.[14] Findings in metabolic processes reveal more information concerning C. perfringens pathogenic nature.[15]
Genome
[edit]Clostridium perfringens has a stable G+C content around 27 to 28 percent and average genome size of 3.5 Mb.[16] Genomes of 56 C. perfringens strains have since been made available on the NCBI genomes database for the scientific research community. Genomic research has revealed surprisingly high diversity in C. perfringens pangenome, with only 12.6 percent core genes, identified as the most divergent Gram-positive bacteria reported.[16] Nevertheless, 16S rRNA regions in between C. perfringens strains are found to be highly conserved (sequence identity >99.1%).[16]
The Clostridium perfringens enterotoxin (CPE)–producing strain has been identified to be a small portion of the overall C. perfringens population (~1-5%) through genomic testing.[17] Advances in genetic information surrounding strain A CPE C. perfringens has allowed techniques such as microbial source tracking (MST) to identify food contamination sources.[17] The CPE gene has been found within chromosomal DNA as well as plasmid DNA. Plasmid DNA has been shown to play and integral role in cell pathogenesis and encodes for major toxins, including CPE.[11]
C. perfringens has been shown to carry plasmid-containing genes for antibiotic resistance. The pCW3 plasmid is the primary conjugation plasmid responsible for creating antibiotic resistance in C. perfringens. Furthermore, the pCW3 plasmid also encodes for multiple toxins found in pathogenic strains of C. perfringens.[18] Antibiotic resistance genes observed thus far include tetracycline resistance, efflux protein, and aminoglycoside resistance.[19]
Within industrial contexts, such as food production, sequencing genomes for pathogenic strains of C. perfringens has become an expanding field of research. Poultry production is impacted directly from this trend as antibiotic-resistant strains of C. perfringens are becoming more common.[12] By performing a meta-genome analysis, researches are capable to identify novel strains of pathogenic bacterium, such as C. perfringens B20.[12]
Motility
[edit]Clostridium perfringens is provisionally identified as non-motile. They lack flagella; however, recent research suggests gliding as a form of motility.[20][21]
Hyper-motile variations
[edit]
In agar plate cultures bacteria with hypermotile variations like SM101 frequently appear around the borders of the colonies. They create long thin filaments that enable them to move quickly, much like bacteria with flagella, according to video imaging of their gliding motion. The causes of the hypermotile phenotype and its immediate descendants were found using genome sequencing. The hypermotile offspring of strains SM101 and SM102, SM124 and SM127, respectively, had 10 and 6 nucleotide polymorphisms (SNPs) in comparison to their parent strains. The hypermotile strains have the common trait of gene mutations related to cell division.[20]
Regulation of gliding motility: The CpAL/VirSR system
[edit]Some strains of C. perfringens cause various diseases like gas gangrene and myonecrosis. Toxins produced that are required for myonecrosis is regulated by the C. perfringens Agr-like (CpAl) system through the VirSR two-component system. The CpAL/VirSR system is a quorum sensing system encoded by other pathogenic clostridia. Myonecrosis starts at the infection site and involves bacteria migrating deeper via gliding motility. Researchers investigated if the CpAL/VirSR system regulates gliding motility. The study demonstrated that the CpAL/VirSR regulates C. perfringens gliding motility. Additionally, the study suggests that gliding bacteria in myonecrosis have increased transcription of toxin genes.[21]
Transformation
[edit]There are two methods of genetic manipulation via experimentation that have been shown to cause genetic transformation in C. perfringens.
Protoplast transformation
[edit]The first report of transformation in C. perfringens involved polyethylene glycol-mediated transformation of protoplasts. The transformation procedure involved the addition of the plasmid DNA to the protoplasts in the presence of high concentrations of polyethylene glycol. During the first protoplast transformation experiment, L-phase variants of C. perfringens were generated by penicillin treatment in the presence 0.4m sucrose. After the transformation procedure was completed, all of the transformed cells were still in the form of L-phase variants. Reversion to vegetative cells was not obtained, but it was observed that autoplasts (protoplasts derived from autolysis) were able to be regenerated to produce rods with cell walls and could be transformed with C. perfringens plasmid DNA.[22]
Electroporation
[edit]Electroporation involves the application of a high-voltage electric field to vegetative bacteria cells for a very short period. This technique resulted in major advances in genetic transformation of C. perfringens, due to the bacteria often displaying itself as a vegetative cell or as dormant spores in food.[23] The electric pulse creates pores in the bacterial cell membrane and allows the passive influx of DNA molecules.[24]
Metabolic processes
[edit]C. perfringens is an aerotolerant anaerobe bacterium that lives in a variety of environments including soil and human intestinal tract.[15] C. perfringens is incapable of synthesizing multiple amino acids due to the lack of genes required for biosynthesis.[15] Instead, the bacterium produces enzymes and toxins to break down host cells and import nutrients from the degrading cell.[15]
C. perfringens has a complete set of enzymes for glycolysis and glycogen metabolism. In the fermentation pathway, pyruvate is converted into acetyl-CoA by pyruvate-ferredoxin oxidoreductase, producing CO2 gas and reduced ferredoxin.[25] Electrons from the reduced ferredoxin are transferred to protons by hydrogenase, resulting in the formation of hydrogen molecules (H2) that are released from the cell along with CO2. Pyruvate is also converted to lactate by lactate dehydrogenase, whereas acetyl-CoA is converted into ethanol, acetate, and butyrate through various enzymatic reactions, completing the anaerobic glycolysis that serves as a potential main energy source for C. perfringens. C. perfringens utilizes a variety of sugars such as fructose, galactose, glycogen, lactose, maltose, mannose, raffinose, starch, and sucrose, and various genes for glycolytic enzymes. The amino acids of these various enzymes and sugar molecules are converted to propionate through propionyl-CoA, which results in energy production.[25]
Virulence
[edit]Membrane-damaging enzymes, pore-forming toxins, intracellular toxins, and hydrolytic enzymes are the functional categories into which C. perfringens' virulence factors may be divided. These virulence factor-encoding genes can be found on chromosomes and large plasmids.[26]
Carbohydrate-active enzymes
[edit]The human gastrointestinal tract is lined with intestinal mucosa that secrete mucus and act as a defense mechanism against pathogens, toxins, and harmful substances. Mucus is made up of mucins containing several O-linked glycan glycoproteins that recognizes and forms a barrier around microbes, preventing them from attaching to endothelial cells and infecting them.[27][28] C. perfringens can secrete different carbohydrate-active enzymes (CAZymes) that aid in degrading mucins and other O-glycans within the intestinal mucosa. These enzymes include: Sialidases, Hexosaminidases, Galactosidases, and Fucosidases belonging to various glycoside hydrolase families.[28]
Sialidase
[edit]Sialidases, also called neuraminidases, function to breakdown mucin by hydrolyzing the terminal sialic acid residues located within the protein through the process of desialylation. C. perfringens has three sialidases belonging to glycoside hydrolase family 33 (GH33): NanH, NanI, and NanJ. All strains of C. perfringens encode for at least one of these enzymes.[28][29]
C. perfringens can secrete NanI and NanJ through secretion signal peptides located on each protein. Research suggests that NanH operates in the cytoplasm of C. perfringens, as it does not contain a secretion signal peptide. NanH contains only a catalytic domain, whereas NanI and NanJ contain a catalytic domain and additional carbohydrate-binding modules (CBMs) to aid in catalytic activity. Located on their N-terminals, NanI contains CBM40, whereas NanJ contains both CBM40 and CBM32. Based on studies analyzing the three-dimensional structure of NanI, its active site has a pocket-like orientation that aids in the removal of sialic acid residues from sialomucins in the intestinal mucosa.[28]
Hexosaminidase
[edit]The mucus layer consists of intestinal mucin glycans, glycolipids, and glycoproteins that contain hexosamines, such as N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc). C. perfringens encodes for eight hexosaminidases that break down hexosamines in the mucus. These hexosaminidases belong to four glycoside hydrolase families: GH36, GH84, GH89, and GH123.[28]
C. perfringens encodes for AagA (CpGH36A) and CpGH36B in glycoside hydrolase family 36 (GH36): AagA removes GalNAc from O-glycans, and CpGH36B is expected to have a similar structure to AagA, but specificities on its function are unknown. NagH, NagI, NagJ, and NagK, belonging to glycoside hydrolase family 84 (GH84), cleave terminal GlcNAc residues using a substrate-assisted digestion mechanism. AgnC (CpGH89), belonging to glycoside hydrolase family 89 (GH89), both cleaves GlcNAc from the ends of mucin glycans and acts on gastric mucin. Belonging to glycoside hydrolase family 123 (GH123), CpNga123 cleaves GalNAc, but research suggests that it only breaks down glycans taken up by C. perfringens due to the absence of a secretion signal peptide.[28]
Galactosidase
[edit]C. perfringens has four galactosidases that belong to the glycoside hydrolase family 2 (GH2): CpGH2A, CpGH2B, CpGH2C, and CpGH2D. Research suggests that these enzymes are effective at breaking down core mucin glycan structures with the ability to bind galactose using CBM51. However, minimal research exists on the specific functioning of galactosidases in C. perfringens.[28]
Fucosidase
[edit]Fucose monosaccharides are located on the terminal ends of core O-linked glycans. C. perfringens encodes for three fucosidases that belong to two glycoside hydrolase families: Afc1 and Afc2 in glycoside hydrolase family 29 (GH29), and Afc3 in glycoside hydrolase family 95 (GH95). Afc3 contains a C-terminal CBM51 and is the only fucosidase that contains a carbohydrate-binding module in C. perfringens. Fucosyl residues tend to cover the ends of glycans and protect them against enzymatic digestion, so research suggests that the ability of fucosidases to cleave complex and diverse fucosyl linkages is due to long-term adaptations in C. perfringens that persisted within close range of mucins.[28]
Major toxins
[edit]There are five major toxins produced by Clostridium perfringens. Alpha, beta, epsilon and enterotoxin are toxins that increase a cells permeability which causes an ion imbalance while iota toxins destroy the cell's actin cytoskeleton.[30] On the basis of which major, "typing" toxins are produced, C. perfringens can be classified into seven "toxinotypes", A, B, C, D, E, F and G:[31]
Toxin Type
|
Alpha | Beta | Epsilon | Iota | Enterotoxin | NetB | Notes |
|---|---|---|---|---|---|---|---|
| A | + | - | - | ||||
| B | + | + | - | - | |||
| C | + | + | - | +/- | - | ||
| D | + | - | + | - | +/- | - | |
| E | + | - | + | +/- | - | ||
| F | + | - | + | - | |||
| G | + | - | + |
Alpha toxin
[edit]Alpha toxin (CPA) is a zinc-containing phospholipase C, composed of two structural domains, which destroy a cell's membrane. Alpha toxins are produced by all five types of C. perfringens. This toxin is linked to gas gangrene of humans and animals. Most cases of gas gangrene has been related to a deep wound being contaminated by soil that harbors C. perfringens.[30][33]
Beta toxin
[edit]Beta toxins (CPB) are a protein that causes hemorrhagic necrotizing enteritis and enterotoxaemia in both animals (type B) and humans (type C) which leads to the infected individual's feces becoming bloody and their intestines necrotizing.[30] Proteolytic enzymes, such as trypsin, can break down CPB, making them ineffective. Therefore, the presence of trypsin inhibitors in colostrum makes CPB especially deadly for mammal offspring.[34]
Epsilon toxin
[edit]Epsilon toxin (ETX) is a protein produced by type B and type D strains of C. perfringens. This toxin is currently ranked the third most potent bacterial toxin known.[35] ETX causes enterotoxaemia in mainly goats and sheep, but cattle are sometime susceptible to it as well. An experiment using mice found that ETX had an LD50 of 50-110 ng/kg.[36] The excessive production of ETX increases the permeability of the intestines. This causes severe edema in organs such as the brain and kidneys.[37]
The very low LD50 of ETX has led to concern that it may be used as a bioweapon. It appeared on the select agent lists of the US CDC and USDA, until it was removed in 2012. There are no human vaccines for this toxin, but effective vaccines for animals exist.[38]
Iota toxin
[edit]Iota toxin (ITX) is a protein produced by type E strains of C. perfringens. Iota toxins are made up of two, unlinked proteins that form a multimeric complex on cells. Iota toxins prevent the formation of filamentous actin. This causes the destruction of the cells cytoskeleton which in turn leads to the death of the cell as it can no longer maintain homeostasis.[39]
Enterotoxin
[edit]This toxin (CPE) causes food poisoning. It alters intracellular claudin tight junctions in gut epithelial cells. This pore-forming toxin also can bind to human ileal and colonic epithelium in vitro and necrotize it. Through the caspase-3 pathway, this toxin can cause apoptosis of affected cells. This toxin is linked to type F strains, but has also been found to be produced by certain types of C, D, and E strains.[40]
Other toxins
[edit]TpeL is a toxin found in type B, C, and G[41] strains. It is in the same protein family as C. difficile toxin A.[42] It does not appear important in the pathogenesis of types B and C infections, but may contribute to virulence in type G strains. It glycosylates Rho and Ras GTPases, disrupting host cell signaling.[41]
Infection
[edit]Tissue necrosis, bacteremia, emphysematous cholecystitis, and gas gangrene, also known as clostridial myonecrosis, have been linked to infections associated with C. perfringens.[43] Research suggests that C. perfringens is capable of engaging in polymicrobial anaerobic infections.[44]
Clostridium perfringens is a common cause of food poisoning in the United States. C. perfringens produces spores, and when these spores are consumed, they produce a toxin that causes diarrhea. Foods cooked in large batches and held at unsafe temperatures (between 40°F and 140°F) are the source of C. perfringens food poisoning outbreaks. Meats such as poultry, beef, and pork are commonly linked to C. perfringens food poisoning.[45] C. perfringens can proliferate in foods that are improperly stored due to the spore's ability to survive normal cooking temperatures. The type A toxin of C. perfringens, also known as the CPA is responsible for food poisoning.[46]
Clostridium perfringens is the most common bacterial agent for gas gangrene.[47] Gas gangrene is induced by α-toxin that embeds itself into the plasma membrane of cells and disrupts normal cellular function by altering membrane structure.[43] Some symptoms include blisters, tachycardia, swelling, and jaundice.[47]
C. perfringens is most commonly known for foodborne illness but can translocate from a gastrointestinal source into the bloodstream which causes bacteremia. C. perfringens bacteremia can lead to toxin-mediated intravascular hemolysis and septic shock.[48] This is rare as it makes up less than 1% of bloodstream isolates but is highly fatal with a reported mortality rate of 27% to 58%.[49]
Clostridium perfringens food poisoning can also lead to another disease known as enteritis necroticans or clostridial necrotizing enteritis, (also known as pigbel); this is caused by C. perfringens type C. This infection is often fatal. Large numbers of C. perfringens grow in the intestines and secrete exotoxin. This exotoxin causes necrosis of the intestines, varying levels of hemorrhaging, and perforation of the intestine. Inflammation usually occurs in sections of the jejunum, midsection of the small intestine.[50] Perfringolysin O (pfoA)-positive C. perfringens strains were also associated with the rapid onset of necrotizing enterocolitis in preterm infants.[51]
A strain of C. perfringens might be implicated in multiple sclerosis (MS) nascent (Pattern III) lesions.[52] Tests in mice found that two strains of intestinal C. perfringens that produced epsilon toxins (ETX) caused MS-like damage in the brain, and earlier work had identified this strain of C. perfringens in a human with MS.[53][54] MS patients were found to be 10 times more immune-reactive to the epsilon toxin than healthy people.[55]
Tissue gas occurs when C. perfringens infects corpses. It causes extremely accelerated decomposition and can only be stopped by embalming the corpse. Tissue gas most commonly occurs to those who have died from gangrene, large decubitus ulcers, necrotizing fasciitis or to those who had soil, feces, or water contaminated with C. perfringens forced into an open wound.[56]
Clinical Manifestations
[edit]Clostridium perfringens infections can lead to various clinical manifestations, ranging from mild gastrointestinal symptoms to life-threatening conditions. The most common presentation is food poisoning, characterized by acute abdominal pain, diarrhea, and, in some cases, vomiting, typically occurring 6 to 24 hours after the ingestion of contaminated food. Unlike many other foodborne illnesses, fever is usually absent. Symptoms are usually self-limiting and resolve within 24 to 48 hours; however, severe dehydration can occur in cases of significant fluid loss. Symptoms of dehydration include dry mouth, decreased urine output, dizziness, and fatigue. Severe symptoms such as diarrhea that persists for more than 48 hours, the inability to keep fluids down, or signs of severe dehydration may necessitate medical attention.[57] Most people are able to recover from C. perfringens food poisoning without treatment. However, people experience diarrhea are usually instructed to drink water or rehydration solutions.[58]
Gas gangrene caused by Clostridium perfringens is characterized by severe symptoms, including intense pain at the injury site, fever, rapid heart rate, sweating, and anxiety. The affected area may show signs of swelling, discoloration (ranging from pale to dark red or purplish), and large, discolored blisters filled with foul-smelling fluid. As the toxins spread, skin and muscle tissue are rapidly destroyed, leading to large areas of dead tissue, gas pockets under the skin (crepitus), and possible renal failure due to red blood cell destruction. Sepsis and septic shock may also occur, which can be fatal.[59]
Necrotizing enteritis caused by Clostridium perfringens presents with a wide range of symptoms, which can vary in severity. The clinical signs range from mild diarrhea to more severe manifestations such as intense abdominal pain, vomiting, bloody stools, and even septic shock. In the most serious cases, the infection can lead to death.[60]
Diagnosis
[edit]The diagnosis of Clostridium perfringens food poisoning relies on laboratory detection of the bacteria or its toxin in either a patient’s stool sample or contaminated food linked to the illness. A positive stool culture would have growth of at least 10 cfu/g of C. perfringens. Stool studies include WBCs, ova, and parasites in order to rule out other potential etiologies. ELISA testing is used to detect the CPA toxin. Diagnosing C. perfringens food poisoning is relatively uncommon for several reasons.[61]Most individuals with this foodborne illness do not seek medical care or submit a stool sample for testing, and routine testing for C. perfringens is not typically performed in clinical laboratories. Additionally, public health laboratories generally conduct testing for this pathogen only in the event of an outbreak.[62]
The diagnosis of gas gangrene typically involves several methods to confirm the infection. Imaging techniques such as X-rays, CT scans, or MRIs can reveal gas bubbles or tissue changes indicative of muscle damage. Additionally, bacterial staining or culture of fluid taken from the wound helps identify Clostridium perfringens and other bacteria responsible for the infection. In some cases, a biopsy is performed, where a sample of the affected tissue is analyzed for signs of damage or necrosis.[59]
The diagnosis of clostridial necrotizing enteritis is primarily based on the patient's clinical symptoms, which can include severe abdominal pain, vomiting, and bloody diarrhea. Additionally, confirmation of the presence of Clostridium perfringens type C toxin in stool samples is crucial for accurate diagnosis.[60]
Epidemiology
[edit]Clostridium perfringens is responsible for an estimated 966,000 cases annually, or about 10.3% of all foodborne illnesses in which a pathogen is identified. Transmission typically occurs when food contaminated with C. perfringens spores is consumed, allowing the bacteria to produce a toxin in the intestines that causes diarrhea. Outbreaks are often associated with foods cooked in large batches, such as poultry, meat, and gravy, and held at unsafe temperatures between 40-140°F, which allows the bacteria to thrive. These outbreaks tend to occur in settings where large groups are served, such as hospitals, school cafeterias, prisons, nursing homes, and catered events. In most cases, C. perfringens infection causes mild symptoms, including watery diarrhea and mild abdominal cramps, with symptoms typically appearing 8 to 12 hours after consuming contaminated food and resolving within 24 hours. About 90% of affected individuals recover without seeking medical attention, usually within two days. However, vulnerable groups such as the elderly, young children, and immunocompromised individuals face a higher risk of severe complications like dehydration, which can lead to more serious illness or, in rare cases, death. Each year, C. perfringens infections result in approximately 438 hospitalizations and 26 deaths, accounting for 0.8% of foodborne illness-related hospitalizations and 1.9% of associated deaths. Outbreaks are most common in November and December, coinciding with holiday foods like turkey and roast beef. The economic burden of C. perfringens is significant, estimated at $342.7 million annually, including $53.2 million in medical costs, $64.3 million in productivity loss, and $225 million related to fatalities.[63][64]
Clostridial necrotizing enteritis is rare in the United States; typically, it occurs in populations with a higher risk. Data show that of the 9.4 million cases of foodborne illness in the United States each year, only about 11% are caused by Clostridium perfringens.[65] "Risk factors for enteritis necroticans include protein-deficient diet, unhygienic food preparation, sporadic feasts of meat (after long periods of a protein-deficient diet), diets containing large amounts of trypsin inhibitors (sweet potatoes), and areas prone to infection of the parasite Ascaris (produces a trypsin inhibitor). This disease is contracted in populations living in New Guinea, parts of Africa, Central America, South America, and Asia.[50]
Risk factors for gas gangrene include severe injuries, abdominal surgeries, and underlying health conditions such as colon cancer, diseases of the blood vessels, diabetes, and diverticulitis. However, the most common way to get gas gangrene is through a traumatic injury. In the United States, there is only about 1000 cases of gas gangrene per year. When addressed with adequate care, gas gangrene has a mortality rate of 20-30% but has a mortality rate of 100% if left untreated.[66]
Food poisoning incidents
[edit]On May 7, 2010, 42 residents and 12 staff members at a Louisiana (USA) state psychiatric hospital were affected and experienced vomiting, abdominal cramps, and diarrhea. Three patients died within 24 hours. The outbreak was linked to chicken which was cooked a day before it was served and was not cooled down according to hospital guidelines. The outbreak affected 31% of the residents of the hospital and 69% of the staff who ate the chicken. How many of the affected residents ate the chicken is unknown.[67]
In May 2011, a man died after allegedly eating food contaminated with the bacteria on a transatlantic American Airlines flight. The man's wife and daughter were suing American and LSG Sky Chefs, the German company that prepared the inflight food.[68]
In December 2012, a 46-year-old woman died two days after eating a Christmas Day meal at a pub in Hornchurch, Essex, England. She was among about 30 people to fall ill after eating the meal. Samples taken from the victims contained C. perfringens. The hotel manager and the cook were jailed for forging cooking records relating to the cooking of the turkey.[69]
In December 2014, 87-year-old Bessie Scott died three days after eating a church potluck supper in Nackawic, New Brunswick, Canada. Over 30 other people reported signs of gastrointestinal illness, diarrhea, and abdominal pain. The province's acting chief medical officer says, Clostridium perfringens is the bacteria [sic] that most likely caused the woman's death.[70]
In October 2016, 66-year-old Alex Zdravich died four days after eating an enchilada, burrito, and taco at Agave Azul in West Lafayette, Indiana, United States. Three others who dined the same day reported signs of foodborne illness, which were consistent with the symptoms and rapid onset of C. perfringens infection. They later tested positive for the presence of the bacteria, but the leftover food brought home by Zdravich tested negative.[71][72]
In November 2016, food contaminated with C. perfringens caused three individuals to die, and another 22 to be sickened, after a Thanksgiving luncheon hosted by a church in Antioch, California, United States.[73]
In January 2017, a mother and her son sued a restaurant in Rochester, New York, United States, as they and 260 other people were sickened after eating foods contaminated with C. perfringens. "Officials from the Monroe County Department of Public Health closed down the Golden Ponds after more than a fourth of its Thanksgiving Day guests became ill. An inspection revealed a walk-in refrigerator with food spills and mold, a damaged gasket preventing the door from closing, and mildew growing inside."[74]
In July 2018, 647 people reported symptoms after eating at a Chipotle Mexican Grill restaurant in Powell, Ohio, United States. Stool samples tested by the CDC tested positive for C. perfringens.[75]
In November 2018, approximately 300 people in Concord, North Carolina, United States, were sickened by food at a church barbecue that tested positive for C. perfringens.[76]
In 2021, dozens of hospital workers in Alaska were sick and it was traced back to a Cubano Sandwich. Health officials wrote that almost all symptoms resolved within 24 hours. No one who ate the food reportedly needed hospitalization. It is rare for Alaska to see an outbreak with this magnitude when it's not associated with some sort of national food borne illness.[77]
Prevention
[edit]Preventing Clostridium perfringens contamination and growth involves careful food handling, proper cooking, and appropriate storage practices. Most foods, especially beef and chicken, can be protected by cooking them to the recommended internal temperatures. Using a kitchen thermometer is the most reliable way to check that meats reach safe cooking temperatures. As a general rule, food should be avoided if it smells, tastes, looks off, or has been left out at unsafe temperatures for a long period of time. [78]
C. perfringens spores can multiply within a temperature range of 59°F (15°C) to 122°F (50°C).[79] To prevent bacterial growth, leftovers should be refrigerated within two hours of preparation, with their temperature chilled down to below 40°F (4°C). Large portions of food that contain meat, should be divided into smaller containers before refrigeration to ensure even cooling. Before serving leftovers, they should be reheated to at least 165°F (74°C) to destroy any bacteria that may have grown during storage.[78]
High-risk foods, such as canned vegetables, smoked or cured meats, and salted or smoked fish, require additional attention. Improper processing or storage can allow bacteria to grow and produce dangerous toxins. Signs of contamination, such as unusual odors, changes in texture, or bulging cans (also known as "bombage"), indicate food spoilage and should be disposed.[80]
Preventing gas gangrene involves taking precautions to avoid bacterial infections. Healthcare providers follow strict protocols to prevent infections, including those caused by Clostridium perfringens. To reduce the risk of gas gangrene, individuals should clean wounds thoroughly with soap and water and seek medical attention for deep or uncleanable wounds. It is also essential to monitor injuries for changes in skin condition or the onset of severe pain. Wearing protective gear when engaging in activities like biking or motorcycling can help prevent injury. Additionally, working with healthcare providers to manage underlying conditions that affect circulation or weaken the immune system can further reduce the risk of infection.[59]
Treatment
[edit]The treatment of Clostridium perfringens infections depends on the type and severity of the condition. For severe infections, such as gas gangrene (clostridial myonecrosis), the primary approach involves surgical debridement of the affected area. This procedure removes devitalized tissue where bacteria grow, which limits the spread of the infection. Antimicrobial therapy is usually started at the same time, with penicillin being the most commonly used drug.[81] However, C. perfringens shows different resistance patterns with about 20% of strains being resistant to clindamycin, and 10% being resistant to metronidazole.[82] C. perfringens is often more susceptible to vancomycin when compared to other pathogenic Clostridia, making it an alternative option for treatment in some cases.[81]
Therapies, such as hyperbaric oxygen therapy (HBOT), may also be used for severe clostridial tissue infections. HBOT increases oxygen delivery to infected tissues, creating an environment that inhibits the growth of anaerobic bacteria like C. perfringens. While not commonly used, HBOT can be beneficial in certain cases.[83]
For foodborne illness caused by C. perfringens, treatment is typically unnecessary. Most people who suffer from food poisoning caused by C. perfringens usually fight off the illness without the need of any antibiotics. Extra fluids should be drank consistently until diarrhea dissipates.[84]
Research
[edit]In recent decades, C. perfringens has shown increasing multidrug resistance, particularly in strains from humans and animals. High resistance levels were found with antibiotics such as tetracycline, erythromycin, and sulfonamides. Genetic factors, misuse of antibiotics, and bacterial evolution are the cause of this issue. This highlights the importance of finding new treatment strategies.[85]
Multilocus Sequence Typing (MLST) and Whole Genome Sequencing (WGS) have been used to find the genetic diversity of C. perfringens. These methods have identified 195 distinct sequence types grouped into 25 clonal complexes from 322 genomes. Phylogenetic groups were also found in multiple different hosts and environmental sources. This highlights the bacteria's transmission potential and adaptability across species.[86]
References
[edit]- ^ Ryan, Kenneth J.; Ray, C. George (2004). Sherris Medical Microbiology : an Introduction to Infectious Diseases (4th ed.). New York: McGraw-Hill. p. 310. ISBN 978-0-8385-8529-0.
- ^ Kiu, R; Hall, L. J. (2018). "An update on the human and animal enteric pathogen Clostridium perfringens". Emerging Microbes & Infections. 7 (141): 141. doi:10.1038/s41426-018-0144-8. PMC 6079034. PMID 30082713.
- ^ "BioNumber Details Page". BioNumbers.
- ^ "Foodborne Illnesses and Germs". Centers for Disease Control and Prevention (CDC). 2018-02-16. Retrieved 18 February 2018.
- ^ Juckett, G; Bardwell, G; McClane, B; Brown, S (2008). "Microbiology of salt rising bread". The West Virginia Medical Journal. 104 (4): 26–7. PMID 18646681.
- ^ Hendrix, Nancy; Mackeen, A.; Weiner, Stuart (2011-01-24). "Clostridium perfringens Sepsis and Fetal Demise after Genetic Amniocentesis". American Journal of Perinatology Reports. 1 (1): 025–028. doi:10.1055/s-0030-1271221. ISSN 2157-6998. PMC 3653538. PMID 23705080.
- ^ Lexicon Orthopaedic Etymology. CRC Press. 1999. p. 128. ISBN 9789057025976.
- ^ Hendrix, Nancy; Mackeen, A.; Weiner, Stuart (2011-01-24). "Clostridium perfringens Sepsis and Fetal Demise after Genetic Amniocentesis". American Journal of Perinatology Reports. 1 (1): 025–028. doi:10.1055/s-0030-1271221. ISSN 2157-6998. PMC 3653538. PMID 23705080.
- ^ Takehara, Masaya; Takagishi, Teruhisa; Seike, Soshi; Ohtani, Kaori; Kobayashi, Keiko; Miyamoto, Kazuaki; Shimizu, Tohru; Nagahama, Masahiro (2016-06-16). "Clostridium perfringens α-Toxin Impairs Innate Immunity via Inhibition of Neutrophil Differentiation". Scientific Reports. 6 (1): 28192. Bibcode:2016NatSR...628192T. doi:10.1038/srep28192. ISSN 2045-2322. PMC 4910053. PMID 27306065.
- ^ Grenda, Tomasz; Jarosz, Aleksandra; Sapała, Magdalena; Grenda, Anna; Patyra, Ewelina; Kwiatek, Krzysztof (2023-05-26). "Clostridium perfringens—Opportunistic Foodborne Pathogen, Its Diversity and Epidemiological Significance". Pathogens. 12 (6): 768. doi:10.3390/pathogens12060768. ISSN 2076-0817. PMC 10304509. PMID 37375458.
- ^ a b Gulliver, Emily L.; Adams, Vicki; Marcelino, Vanessa Rossetto; Gould, Jodee; Rutten, Emily L.; Powell, David R.; Young, Remy B.; D’Adamo, Gemma L.; Hemphill, Jamia; Solari, Sean M.; Revitt-Mills, Sarah A.; Munn, Samantha; Jirapanjawat, Thanavit; Greening, Chris; Boer, Jennifer C. (2023-04-20). "Extensive genome analysis identifies novel plasmid families in Clostridium perfringens". Microbial Genomics. 9 (4). doi:10.1099/mgen.0.000995. ISSN 2057-5858. PMC 10210947. PMID 37079454. S2CID 258238878.
- ^ a b c d Elnar, Arxel G.; Kim, Geun-Bae (2021-11-30). "Complete genome sequence of Clostridium perfringens B20, a bacteriocin-producing pathogen". Journal of Animal Science and Technology. 63 (6): 1468–1472. doi:10.5187/jast.2021.e113. ISSN 2672-0191. PMC 8672250. PMID 34957460.
- ^ Revitt-Mills, Sarah A; Rood, Julian I; Adams, Vicki (2015). "Clostridium perfringens extracellular toxins and enzymes: 20 and counting". Microbiology Australia. 36 (3): 114. doi:10.1071/MA15039. ISSN 1324-4272.
- ^ Wambui, Joseph; Cernela, Nicole; Stevens, Marc J. A.; Stephan, Roger (2021-09-13). "Whole Genome Sequence-Based Identification of Clostridium estertheticum Complex Strains Supports the Need for Taxonomic Reclassification Within the Species Clostridium estertheticum". Frontiers in Microbiology. 12. doi:10.3389/fmicb.2021.727022. ISSN 1664-302X. PMC 8473909. PMID 34589074.
- ^ a b c d Ohtani, Kaori; Shimizu, Tohru (2016-07-05). "Regulation of Toxin Production in Clostridium perfringens". Toxins. 8 (7): 207. doi:10.3390/toxins8070207. ISSN 2072-6651. PMC 4963840. PMID 27399773.
- ^ a b c Kiu, Raymond; Caim, Shabhonam; Alexander, Sarah; Pachori, Purnima; Hall, Lindsay J. (2017). "Probing Genomic Aspects of the Multi-Host Pathogen Clostridium perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors". Frontiers in Microbiology. 8: 2485. doi:10.3389/fmicb.2017.02485. PMC 5733095. PMID 29312194.
- ^ a b Miyamoto, Kazuaki; Li, Jihong; McClane, Bruce A. (2012). "Enterotoxigenic Clostridium perfringens: Detection and Identification". Microbes and Environments. 27 (4): 343–349. doi:10.1264/jsme2.ME12002. ISSN 1342-6311. PMC 4103540. PMID 22504431. S2CID 7743606.
- ^ Adams, Vicki; Han, Xiaoyan; Lyras, Dena; Rood, Julian I. (September 2018). "Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens". Plasmid. 99: 32–39. doi:10.1016/j.plasmid.2018.07.002. PMID 30055188. S2CID 51866356.
- ^ Kiu, Raymond; Caim, Shabhonam; Alexander, Sarah; Pachori, Purnima; Hall, Lindsay J. (2017-12-12). "Probing Genomic Aspects of the Multi-Host Pathogen Clostridium perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors". Frontiers in Microbiology. 8: 2485. doi:10.3389/fmicb.2017.02485. ISSN 1664-302X. PMC 5733095. PMID 29312194.
- ^ a b Liu, Hualan; McCord, Kristin D.; Howarth, Jonathon; Popham, David L.; Jensen, Roderick V.; Melville, Stephen B. (July 2014). "Hypermotility in Clostridium perfringens Strain SM101 Is Due to Spontaneous Mutations in Genes Linked to Cell Division". Journal of Bacteriology. 196 (13): 2405–2412. doi:10.1128/JB.01614-14. ISSN 0021-9193. PMC 4054169. PMID 24748614.
- ^ a b Valeriani, Renzo G.; Beard, LaMonta L.; Moller, Abraham; Ohtani, Kaori; Vidal, Jorge E. (2020-12-01). "Gas gangrene-associated gliding motility is regulated by the Clostridium perfringens CpAL/VirSR system". Anaerobe. 66: 102287. doi:10.1016/j.anaerobe.2020.102287. ISSN 1075-9964. PMID 33130105.
- ^ Rood, J I; Cole, S T (December 1991). "Molecular genetics and pathogenesis of Clostridium perfringens". Microbiological Reviews. 55 (4): 621–648. doi:10.1128/mr.55.4.621-648.1991. ISSN 0146-0749. PMC 372840. PMID 1779929.
- ^ Golden, Neal J.; Crouch, Edmund A.; Latimer, Heejeong; Kadry, Abdel-Razak; Kause, Janell (July 2009). "Risk Assessment for Clostridium perfringens in Ready-to-Eat and Partially Cooked Meat and Poultry Products". Journal of Food Protection. 72 (7): 1376–1384. doi:10.4315/0362-028x-72.7.1376. ISSN 0362-028X. PMID 19681258.
- ^ Rood, J I; Cole, S T (1991). "Molecular genetics and pathogenesis of Clostridium perfringens". Microbiological Reviews. 55 (4): 621–648. doi:10.1128/MMBR.55.4.621-648.1991. ISSN 0146-0749. PMC 372840. PMID 1779929.
- ^ a b Shimizu, Tohru; Ohtani, Kaori; Hirakawa, Hideki; Ohshima, Kenshiro; Yamashita, Atsushi; Shiba, Tadayoshi; Ogasawara, Naotake; Hattori, Masahira; Kuhara, Satoru; Hayashi, Hideo (2002-01-22). "Complete genome sequence of Clostridium perfringens , an anaerobic flesh-eater". Proceedings of the National Academy of Sciences. 99 (2): 996–1001. Bibcode:2002PNAS...99..996S. doi:10.1073/pnas.022493799. ISSN 0027-8424. PMC 117419. PMID 11792842.
- ^ Revitt-Mills, Sarah A; Rood, Julian I; Adams, Vicki (2015). "Clostridium perfringens extracellular toxins and enzymes: 20 and counting". Microbiology Australia. 36 (3): 114. doi:10.1071/MA15039. ISSN 1324-4272.
- ^ Ba, Xuli; Jin, Youshun; Ning, Xuan; Gao, Yidan; Li, Wei; Li, Yunhui; Wang, Yihan; Zhou, Jizhang (2024-08-07). "Clostridium perfringens in the Intestine: Innocent Bystander or Serious Threat?". Microorganisms. 12 (8): 1610. doi:10.3390/microorganisms12081610. ISSN 2076-2607. PMC 11356505. PMID 39203452.
- ^ a b c d e f g h Low, Kristin E; Smith, Steven P; Abbott, D Wade; Boraston, Alisdair B (2020-05-30). "The glycoconjugate-degrading enzymes of Clostridium perfringens: Tailored catalysts for breaching the intestinal mucus barrier". Glycobiology. 31 (6): 681–690. doi:10.1093/glycob/cwaa050. ISSN 1460-2423. PMID 32472136.
- ^ Medley, Brendon J.; Low, Kristin E.; Irungu, Jackline D. W.; Kipchumba, Linus; Daneshgar, Parandis; Liu, Lin; Garber, Jolene M.; Klassen, Leeann; Inglis, G. Douglas; Boons, Geert-Jan; Zandberg, Wesley F.; Abbott, D. Wade; Boraston, Alisdair B. (2024-10-01). "A "terminal" case of glycan catabolism: Structural and enzymatic characterization of the sialidases of Clostridium perfringens". Journal of Biological Chemistry. 300 (10): 107750. Bibcode:2024JBiCh.300j7750M. doi:10.1016/j.jbc.2024.107750. ISSN 0021-9258. PMC 11525138. PMID 39251137.
- ^ a b c Stiles, Bradley G.; Barth, Gillian; Barth, Holger; Popoff, Michel R. (2013-11-12). "Clostridium perfringens Epsilon Toxin: A Malevolent Molecule for Animals and Man?". Toxins. 5 (11): 2138–2160. doi:10.3390/toxins5112138. ISSN 2072-6651. PMC 3847718. PMID 24284826.
- ^ a b Johnston MD, Whiteside TE, Allen ME, Kurtz DM (February 2022). "Toxigenic Profile of Clostridium perfringens Strains Isolated from Natural Ingredient Laboratory Animal Diets". Comparative Medicine. 72 (1): 50–58. doi:10.30802/AALAS-CM-22-000013. PMC 8915413. PMID 35148812.
- ^ Rood, Julian I.; Adams, Vicki; Lacey, Jake; Lyras, Dena; McClane, Bruce A.; Melville, Stephen B.; Moore, Robert J.; Popoff, Michel R.; Sarker, Mahfuzur R.; Songer, J. Glenn; Uzal, Francisco A.; Van Immerseel, Filip (2018-10-01). "Expansion of the Clostridium perfringens toxin-based typing scheme". Anaerobe. 53: 5–10. doi:10.1016/j.anaerobe.2018.04.011. ISSN 1075-9964. PMC 6195859. PMID 29866424.
- ^ Li, Ming; Li, Ning (2021-06-16). "Clostridium perfringens bloodstream infection secondary to acute pancreatitis: A case report". World Journal of Clinical Cases. 9 (17): 4357–4364. doi:10.12998/wjcc.v9.i17.4357. ISSN 2307-8960. PMC 8173429. PMID 34141801.
- ^ Garcia, J.P.; Beingesser, J.; Fisher, D.J.; Sayeed, S.; McClane, B.A.; Posthaus, H.; Uzal, F.A. (4 January 2012). "The effect of Clostridium perfringens type C strain CN3685 and its isogenic beta toxin null mutant in goats". Veterinary Microbiology. 157 (3–4): 412–419. doi:10.1016/j.vetmic.2012.01.005. PMC 3348370. PMID 22296994.
- ^ Alves, Guilherme Guerra; Machado de Ávila, Ricardo Andrez; Chávez-Olórtegui, Carlos Delfin; Lobato, Francisco Carlos Faria (2014-12-01). "Clostridium perfringens epsilon toxin: The third most potent bacterial toxin known". Anaerobe. 30: 102–107. doi:10.1016/j.anaerobe.2014.08.016. ISSN 1075-9964. PMID 25234332.
- ^ Xin, Wenwen; Wang, Jinglin (2019-09-01). "Clostridium perfringens epsilon toxin: Toxic effects and mechanisms of action". Biosafety and Health. 1 (2): 71–75. doi:10.1016/j.bsheal.2019.09.004. ISSN 2590-0536. S2CID 208690896.
- ^ Geng, Zhijun; Kang, Lin; Huang, Jing; Gao, Shan; Wang, Jing; Yuan, Yuan; Li, Yanwei; Wang, Jinglin; Xin, Wenwen (2021-07-30). "Epsilon toxin from Clostridium perfringens induces toxic effects on skin tissues and HaCaT and human epidermal keratinocytes". Toxicon. 198: 102–110. Bibcode:2021Txcn..198..102G. doi:10.1016/j.toxicon.2021.05.002. ISSN 0041-0101. PMID 33965432. S2CID 234343237.
- ^ Stiles, Bradley G.; Barth, Gillian; Popoff, Michel R. P (2018). "Clostridium Perfringens Epsilon Toxin". Medical Aspects of Biological Warfare (2 ed.). Health Readiness Center of Excellence (US Army). ISBN 9780160941597.
- ^ Sakurai, Jun; Nagahama, Masahiro; Oda, Masataka; Tsuge, Hideaki; Kobayashi, Keiko (2009-12-23). "Clostridium perfringens Iota-Toxin: Structure and Function". Toxins. 1 (2): 208–228. doi:10.3390/toxins1020208. ISSN 2072-6651. PMC 3202787. PMID 22069542.
- ^ Kiu, Raymond; Hall, Lindsay J. (2018-12-01). "An update on the human and animal enteric pathogen Clostridium perfringens". Emerging Microbes & Infections. 7 (1): 141. doi:10.1038/s41426-018-0144-8. ISSN 2222-1751. PMC 6079034. PMID 30082713.
- ^ a b Orrell, KE; Melnyk, RA (18 August 2021). "Large Clostridial Toxins: Mechanisms and Roles in Disease". Microbiology and Molecular Biology Reviews. 85 (3): e0006421. doi:10.1128/MMBR.00064-21. PMC 8483668. PMID 34076506.
- ^ Chen, J; McClane, BA (June 2015). "Characterization of Clostridium perfringens TpeL toxin gene carriage, production, cytotoxic contributions, and trypsin sensitivity". Infection and Immunity. 83 (6): 2369–81. doi:10.1128/IAI.03136-14. PMC 4432761. PMID 25824828.
- ^ a b Hendrix, Nancy; Mackeen, A.; Weiner, Stuart (2011-01-24). "Clostridium perfringens Sepsis and Fetal Demise after Genetic Amniocentesis". American Journal of Perinatology Reports. 1 (1): 025–028. doi:10.1055/s-0030-1271221. ISSN 2157-6998. PMC 3653538. PMID 23705080.
- ^ Takehara, Masaya; Takagishi, Teruhisa; Seike, Soshi; Ohtani, Kaori; Kobayashi, Keiko; Miyamoto, Kazuaki; Shimizu, Tohru; Nagahama, Masahiro (2016-06-16). "Clostridium perfringens α-Toxin Impairs Innate Immunity via Inhibition of Neutrophil Differentiation". Scientific Reports. 6 (1): 28192. Bibcode:2016NatSR...628192T. doi:10.1038/srep28192. ISSN 2045-2322. PMC 4910053. PMID 27306065.
- ^ CDC (2024-05-16). "About C. perfringens food poisoning". Clostridium perfringens Food Poisoning. Retrieved 2024-11-20.
- ^ Yao, Phil Y.; Annamaraju, Pavan (2024), "Clostridium perfringens Infection", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 32644475, retrieved 2024-11-20
- ^ a b "Gas gangrene: MedlinePlus Medical Encyclopedia".
- ^ "Cytarabine". Reactions Weekly. 1959 (1): 223. 2023-06-03. doi:10.1007/s40278-023-40395-7. ISSN 1179-2051. S2CID 259027022.
- ^ Millard, Michael A.; McManus, Kathleen A.; Wispelwey, Brian (2016). "Severe Sepsis due to Clostridium perfringens Bacteremia of Urinary Origin: A Case Report and Systematic Review". Case Reports in Infectious Diseases. 2016: 1–5. doi:10.1155/2016/2981729. ISSN 2090-6625. PMC 4779822. PMID 26998370.
- ^ a b Lentino, Joseph R. (2016-01-01). "Clostridial Necrotizing Enteritis". Merck Manuel. Merck Sharp & Dohme Corp. Retrieved 2016-09-27.
- ^ Kiu, Raymond; Shaw, Alexander G.; Sim, Kathleen; Acuna-Gonzalez, Antia; Price, Christopher A.; Bedwell, Harley; Dreger, Sally A.; Fowler, Wesley J.; Cornwell, Emma; Pickard, Derek; Belteki, Gusztav; Malsom, Jennifer; Phillips, Sarah; Young, Gregory R.; Schofield, Zoe (June 2023). "Particular genomic and virulence traits associated with preterm infant-derived toxigenic Clostridium perfringens strains". Nature Microbiology. 8 (6): 1160–1175. doi:10.1038/s41564-023-01385-z. ISSN 2058-5276. PMC 10234813. PMID 37231089.
- ^ Rumah, Kareem Rashid; Linden, Jennifer; Fischetti, Vincent A.; Vartanian, Timothy; Esteban, Francisco J. (16 October 2013). "Isolation of Clostridium perfringens Type B in an Individual at First Clinical Presentation of Multiple Sclerosis Provides Clues for Environmental Triggers of the Disease". PLOS ONE. 8 (10): e76359. Bibcode:2013PLoSO...876359R. doi:10.1371/journal.pone.0076359. PMC 3797790. PMID 24146858.
- ^ "Multiple sclerosis 'linked to food bug'". BBC. 29 January 2014. Retrieved 29 January 2014.
- ^ Reder, Anthony T. (2023-05-01). "Clostridium epsilon toxin is excessive in multiple sclerosis and provokes multifocal lesions in mouse models". Journal of Clinical Investigation. 133 (9). doi:10.1172/JCI169643. ISSN 1558-8238. PMC 10145922. PMID 37115699. S2CID 258375399.
- ^ Woerner, Amanda (29 January 2014). "Bacterial toxin may trigger multiple sclerosis, research finds". Fox News.
- ^ "Clostridium perfringens" (PDF). ldh.la.gov/. Louisiana Office of Public Health. Retrieved 14 June 2024.
- ^ "Symptoms of C. perfringens food poisoning". Centers for Disease Control and Prevention. 13 May 2024. Retrieved 12 November 2024.
- ^ CDC (2024-05-13). "Treating C. perfringens food poisoning". Clostridium perfringens Food Poisoning. Retrieved 2024-11-20.
- ^ a b c "Gas Gangrene". Cleveland Clinic. February 2024.
- ^ a b "Clostridial Necrotizing Enteritis - Clostridial Necrotizing Enteritis". MSD Manual Professional Edition. Retrieved 2024-11-21.
- ^ Yao, Phil Y.; Annamaraju, Pavan (2024), "Clostridium perfringens Infection", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 32644475, retrieved 2024-11-21
- ^ CDC (2024-05-13). "Diagnosing C. perfringens food poisoning". Clostridium perfringens Food Poisoning. Retrieved 2024-11-21.
- ^ Hoffmann, Sandra; Maculloch, Bryan; Batz, Michael. "Economic Burden of Major Foodborne Illnesses Acquired in the United States". www.ers.usda.gov. Retrieved 2024-11-21.
- ^ CDC (2024-05-16). "About C. perfringens food poisoning". Clostridium perfringens Food Poisoning. Retrieved 2024-11-21.
- ^ Scallan, Elaine; Hoekstra, Robert M.; Angulo, Frederick J.; Tauxe, Robert V.; Widdowson, Marc-Alain; Roy, Sharon L.; Jones, Jeffery L.; Griffin, Patricia M. (January 2011). "Foodborne Illness Acquired in the United States—Major Pathogens". Emerging Infectious Diseases. 17 (1): 7–15. doi:10.3201/eid1701.P11101. PMC 3375761. PMID 21192848.
- ^ Buboltz, Jerome B.; Murphy-Lavoie, Heather M. (2024), "Gas Gangrene", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 30725715, retrieved 2024-11-21
- ^ "Fatal Foodborne Clostridium perfringens Illness at a State Psychiatric Hospital — Louisiana, 2010". Centers for Disease Control and Prevention. Retrieved 16 November 2013.
- ^ Mohn, Tanya (1 December 2011). "Passenger dies in-flight, family says airline to blame". Overhead Bin. MSNBC. Retrieved 2012-02-13.
- ^ "Pub chef and manager jailed over Christmas dinner that left a diner dead". The Guardian. 23 January 2015. Retrieved 3 August 2015.
- ^ "Woman's death likely caused by bacteria from Christmas supper". CBC. 12 December 2014.
- ^ "Food poisoning death at Indiana restaurant kept secret for months". 13 WTHR Indianapolis. 2017-07-17. Archived from the original on 2017-07-20. Retrieved 2017-07-18.
- ^ (WTHR), Susan Batt. "Agave Azul Tippecanoe Co Food Poisoning Finding Summary". www.documentcloud.org. Retrieved 2017-07-18.
- ^ "Bacteria that killed 3 at Antioch Thanksgiving dinner pinpointed". SFGate. Retrieved 2016-12-20.
- ^ "Mother, son sue eatery for Thanksgiving dinner food poisoning - Food Safety News". 6 January 2017.
- ^ "CDC releases test findings after hundreds sickened at Powell Chipotle - Columbus Dispatch". 16 August 2018. Archived from the original on 16 August 2018. Retrieved 16 August 2018.
- ^ "Strain of food poisoning causes illness at North Carolina church barbecue". November 2018.
- ^ "Cubano Sandwiches with Clostridium Perfringens Found in Alaska Investigation". Food Safety News. August 12, 2021.
- ^ a b CDC (2024-05-16). "Preventing C. perfringens Food Poisoning". Clostridium perfringens Food Poisoning. Retrieved 2024-11-21.
- ^ Taormina, Peter J.; Dorsa, Warren J. (2004-07-01). "Growth Potential of Clostridium perfringens during Cooling of Cooked Meats". Journal of Food Protection. 67 (7): 1537–1547. doi:10.4315/0362-028X-67.7.1537. ISSN 0362-028X. PMID 15270517.
- ^ Bilska, Agnieszka; Wochna, Krystian; Habiera, Małgorzata; Serwańska-Leja, Katarzyna (2024-08-18). "Health Hazard Associated with the Presence of Clostridium Bacteria in Food Products". Foods. 13 (16): 2578. doi:10.3390/foods13162578. ISSN 2304-8158. PMC 11353352. PMID 39200505.
- ^ a b Di Bella, Stefano; Antonello, Roberta Maria; Sanson, Gianfranco; Maraolo, Alberto Enrico; Giacobbe, Daniele Roberto; Sepulcri, Chiara; Ambretti, Simone; Aschbacher, Richard; Bartolini, Laura; Bernardo, Mariano; Bielli, Alessandra; Busetti, Marina; Carcione, Davide; Camarlinghi, Giulio; Carretto, Edoardo (June 2022). "Anaerobic bloodstream infections in Italy (ITANAEROBY): A 5-year retrospective nationwide survey". Anaerobe. 75: 102583. doi:10.1016/j.anaerobe.2022.102583. hdl:11368/3020691. PMID 35568274.
- ^ Geremia, Nicholas; Sanson, Gianfranco; Principe, Luigi; Antonello, Roberta Maria; Zerbato, Verena; Luzzati, Roberto; Maraolo, Alberto Enrico; Giacobbe, Daniele Roberto; Sepulcri, Chiara; Ambretti, Simone; Aschbacher, Richard; Bartolini, Laura; Bernardo, Mariano; Bielli, Alessandra; Busetti, Marina (August 2024). "A subanalysis of Clostridium perfringens bloodstream infections from a 5-year retrospective nationwide survey (ITANAEROBY)". Anaerobe. 90: 102901. doi:10.1016/j.anaerobe.2024.102901. PMID 39214165.
- ^ Gibson, A.; Davis, F. M. (1986-08-27). "Hyperbaric oxygen therapy in the management of Clostridium perfringens infections". The New Zealand Medical Journal. 99 (808): 617–620. ISSN 0028-8446. PMID 3462561.
- ^ CDC (2023-03-24). "Prevent Illness From C. perfringens". Centers for Disease Control and Prevention. Retrieved 2023-10-01.
- ^ Ba, Xuli; Jin, Youshun; Ning, Xuan; Gao, Yidan; Li, Wei; Li, Yunhui; Wang, Yihan; Zhou, Jizhang (August 2024). "Clostridium perfringens in the Intestine: Innocent Bystander or Serious Threat?". Microorganisms. 12 (8): 1610. doi:10.3390/microorganisms12081610. ISSN 2076-2607. PMC 11356505. PMID 39203452.
- ^ Camargo, Anny; Guerrero-Araya, Enzo; Castañeda, Sergio; Vega, Laura; Cardenas-Alvarez, María X.; Rodríguez, César; Paredes-Sabja, Daniel; Ramírez, Juan David; Muñoz, Marina (2022-07-22). "Intra-species diversity of Clostridium perfringens: A diverse genetic repertoire reveals its pathogenic potential". Frontiers in Microbiology. 13. doi:10.3389/fmicb.2022.952081. ISSN 1664-302X. PMC 9354469. PMID 35935202.
External links
[edit]- Clostridium perfringens genomes and related information at PATRIC, a Bioinformatics Resource Center funded by NIAID
- Pathema-Clostridium Resource
- Type strain of Clostridium perfringens at BacDive - the Bacterial Diversity Metadatabase
